Family: Asfarviridae
Covadonga Alonso, Manuel Borca, Linda Dixon, Yolanda Revilla, Fernando Rodriguez and Jose M. Escribano
The citation for this ICTV Report chapter is the summary published as Alonso et al., (2018):
ICTV Virus Taxonomy Profile: Asfarviridae, Journal of General Virology, 99: 613–614.
Corresponding author: Covadonga Alonso ([email protected])
Edited by: Balázs Harrach and Andrew J. Davison
Posted: March 2018
PDF: ICTV_Asfarviridae.pdf
Summary
The family Asfarviridae includes the single species Asfivirus haemorrhagiae, isolates of which have linear dsDNA genomes of 170–194 kbp (Table 1 Asfaviridae). Virions have an internal core, an internal lipid layer, an icosahedral capsid and an outer lipid envelope. Infection of domestic pigs and wild boar results in haemorrhagic fever and, for virulent isolates, death. Infection can be transmitted by contact, ingestion, or by ticks of the genus Ornithodoros. The virus is endemic in Africa where warthogs and bush pigs act as reservoirs. The virus spread from West Africa to the Iberian peninsula, remaining endemic for 30 years, and was finally eradicated from Europe except for the island of Sardinia, Italy. In 2007, the virus again spread out of Africa through the Caucasus to Europe and is currently causing outbreaks in the Russian Federation and several neighbouring countries such as the Baltic Republics, Poland, Czech Republic and Romania.
Table 1. Asfarviridae. Characteristics of members of the family Asfarviridae.
| Characteristic | Description |
| Typical member | African swine fever virus BA71V (U18466), species Asfivirus haemorrhagiae |
| Virion | Multiple layers of core, internal envelope, capsid and external envelope. Polyprotein processing by a viral protease yields multiple subunit structural proteins |
| Genome | Linear dsDNA, 170–194 kbp with complementary terminal loops |
| Replication | Cytoplasmic with an early nuclear phase not fully characterised. Head-to-head concatemer replicative intermediates similar to poxviruses. Transcription and RNA processing uses virus-encoded enzymes |
| Translation | From mRNAs with 5′-caps and 3′-polyadenylation |
| Host range | Domestic pig, wild boar, warthog and bush pig; transmitted by contact, ingestion, or by Ornithodoros ticks |
| Taxonomy | Realm Varidnaviria, kingdom Bamfordvirae, phylum Nucleocytoviricota, class Pokkesviricetes, order Asfuvirales: single species in the single genus Asfivirus |
Virion
Morphology
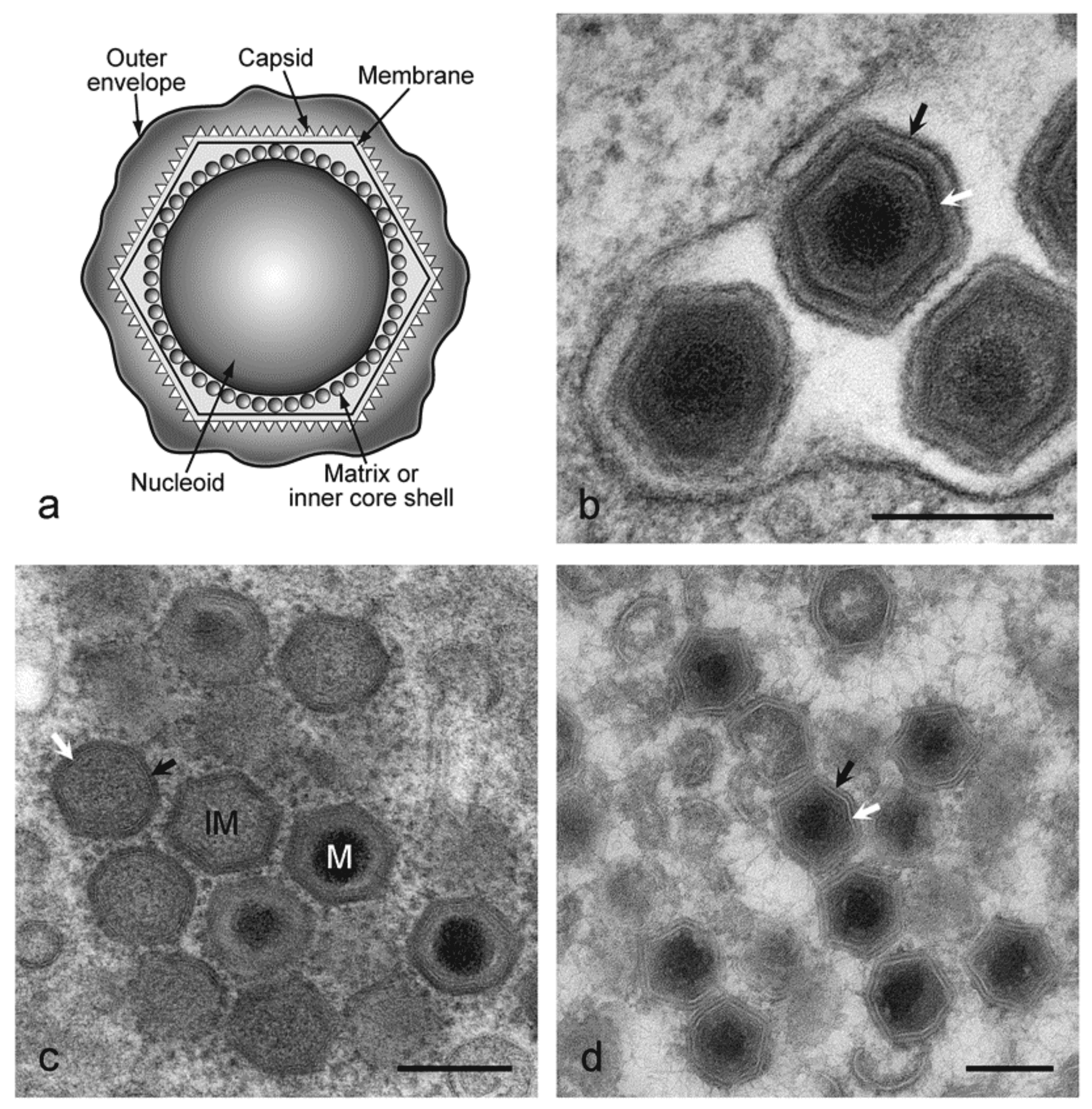
African swine fever virus (ASFV) virions consist of a nucleoprotein core structure, 70–100 nm in diameter, surrounded by an internal lipid layer and an icosahedral capsid, 170–190 nm in diameter, which in turn is eventually surrounded by an external lipid-containing envelope. This external envelope is dispensable for infection. The capsid exhibits icosahedral symmetry (T=189–217) corresponding to 1892–2172 capsomers. Each capsomer is 13 nm in diameter and appears as a hexagonal prism with a central hole; intercapsomeric distance is 7.4–8.1 nm. An internal membrane surrounds the core shell coating the nucleic acid-containing nucleoid (Salas and Andrés 2013). Extracellular enveloped virions have a diameter of 175–215 nm (Figure 1 Asfaviridae).
Physicochemical and physical properties
Virion buoyant density is 1.095 g cm−3 in Percoll, 1.19–1.24 g cm−3 in CsCl; S20,W is about 3500S. Virions are sensitive to ether, chloroform and deoxycholate, and are inactivated at 60 °C within 30 min but survive for years at 20 °C or 4 °C. Infectivity is stable over a wide pH range. Some infectious virus may survive treatment at pH4 or pH13. Infectivity is destroyed by some disinfectants (1% formaldehyde for 6 d, 2% NaOH for 1 d); paraphenylphenolic disinfectants are very effective. Virus is sensitive to irradiation.
Nucleic acid
The genome consists of a single molecule of linear, covalently close-ended, dsDNA of 170–194 kbp. The end sequences are present as two flip-flop forms that are inverted and complementary with respect to each other, and adjacent to both termini are identical arrays of directly repeated 2.1 kbp units. The complete nucleotide sequences of 18 isolates have been determined. These include the tissue culture-adapted Ba71V isolate (ASFV-BA71V) and 17 field isolates from Europe and Africa (Chapman et al., 2008, de Villiers et al., 2010, Portugal et al., 2015, Chapman et al., 2011).
Proteins
Virions contain more than 50 proteins, including a number of enzymes and factors needed for early mRNA transcription and processing (Table 2 Asfarviridae). Enzymes packaged into virions include the multi-subunit RNA polymerase, polyA polymerase, guanylyl transferase and protein kinase. The inhibitor of apoptosis (IAP) homolog protein is also packaged in virions. Virion structural proteins characterized include: on the outer envelope, the CD2v protein (EP402R), P24 (pKP177R), pP2 (pO61R); on the capsid shell, the major capsid protein P72 (pB646L), P49 (pB438L) and pE120R; in the internal envelope, P17 (pD117L), P54 or j13L (pE183L), and probably j18L (pE199L) and j5R (pH108R). The products of a 220 kDa protein (pCP2475L) that is cleaved to give four structural proteins (P150, P37, P14 and P34), and the products of a 62 kDa protein (pCP530R) that is cleaved to give two structural proteins (P35 and P15), are localized in the matrix or inner core shell (Suarez et al., 2010). A virus-encoded protease related to the SUMO-1-specific protease family is involved in cleavage of these polyproteins. Two DNA binding proteins, P10 (pA78R) and P14.5 (pE120R), are present in virions. The virus encodes components of a redox pathway, including the pB119L (or 9GL), pE248R and pA151R proteins, of which the pA151R and pB119L proteins are non-structural. The pB602L protein is a chaperone required for assembly of the P72 capsid protein into virions (Cobbold et al., 2001).
Table 2 Asfarviridae. Functions of African swine fever virus proteins.
| Gene function | Gene name | Predicted protein mass (kDa) |
| Nucleotide metabolism, transcription, replication and repair | ||
| Thymidylate kinase | A240L | 27.8 |
| Thymidine kinase | K196R | 22.4 |
| Deoxyuridine triphosphatase* | E165R | 18.3 |
| Ribonucleotide reductase (small subunit) | F334L | 39.8 |
| Ribonucleotide reductase (large subunit) | F778R | 87.5 |
| DNA polymerase α-like | G1211R | 139.8 |
| DNA topoisomerase type II* | P1192R | 135.5 |
| Proliferating cell nuclear antigen (PCNA)-like | E301R | 35.3 |
| DNA polymerase X-like* | O174L | 20.3 |
| DNA ligase* | NP419L | 48.2 |
| AP endonuclease class II* | E296R | 33.5 |
| RNA polymerase subunit 2 | EP1242L | 139.9 |
| RNA polymerase subunit 6 | C147L | 16.7 |
| RNA polymerase subunit 1 | NP1450L | 163.7 |
| RNA polymerase subunit 3 | H359L | 41.3 |
| RNA polymerase subunit 5 | D205R | 23.7 |
| RNA polymerase subunit 7 | D339L | 39 |
| RNA polymerase subunit 10 | CP80R | 9.0 |
| Transcription factor IIB-like | C315R | 37.0 |
| Helicase superfamily II | A859L | 27.8 |
| Helicase superfamily II | F1055L | 123.9 |
| Helicase superfamily II | B962L | 109.6 |
| Helicase superfamily II | D1133L | 129.3 |
| Helicase superfamily II | Q706L | 80.4 |
| Helicase superfamily II | QP509L | 58.1 |
| Transcription factor SII | I243L | 28.6 |
| Guanylyl transferase* | NP868R | 29.9 |
| PolyA polymerase large subunit | C475L | 54.8 |
| FTSJ-like methyl transferase domain** | EP424R | 49.3 |
| ERCC4 nuclease domain** | EP364R | 40.9 |
| Lambda-like exonuclease | D345L | 39.4 |
| VV A2L-like transcription factor** | B385R | 45.3 |
| VV A7L-like transcription factor** | G1340L | 155.0 |
| VV VLTF2-like late transcription factor** | B175L | 20.3 |
| FCS-like finger DNA primase | C962R | 111.3 |
| Other enzymes | ||
| Prenyltransferase* | B318L | 35.9 |
| Serine protein kinase* | R298L | 35.1 |
| Ubiquitin conjugating enzyme* | I215L | 24.7 |
| Decapping enzyme (g5R)* | D250R | 29.9 |
| Host cell interactions | ||
| IAP apoptosis inhibitor* | A224L | 26.6 |
| Bcl-2 apoptosis inhibitor* | A179L | 21.1 |
| Inhibitor of host gene transcription* | A238L | 28.2 |
| C-type lectin-like* | EP153R | 18.0 |
| Similar to HSV ICP34.5 neurovirulence factor** | DP71L | 8.5 |
| Nif S-like | QP383R | 42.5 |
| ERV 1-like. Involved in redox metabolism*,** | B119L | 14.4 |
| Phosphoprotein binds to ribonucleoprotein-K | CP204L | 30.0 |
| UK virulence factor | DP96R | 10.7 |
| Structural proteins and proteins involved in morphogenesis | ||
| P22 | KP177R | 20.2 |
| Histone-like | A104R | 11.5 |
| P11.5 | A137R | 21.1 |
| P10 | K78R | 8.4 |
| pA151R. Contains CXXC motif similar to that in thioredoxins. Binds to E248R protein. Not incorporated into virions. Possible component of redox pathway. | A151R | 17.5 |
| P72 major capsid protein. Involved in virus entry | B646L | 73.2 |
| P49. Required for formation of vertices in icosahedral capsid | B438L | 49.3 |
| Chaperone. Involved in folding of capsid. Not incorporated into virions | B602L | 45.3 |
| SUMO-1-like protease. Involved in polyprotein cleavage | S273R | 31.6 |
| pp220 polyprotein precursor of p150, p37, p14 and p34. Required for packaging of nucleoprotein core | CP2475L | 281.5 |
| P32 (P30) phosphoprotein. Involved in virus entry | CP204L | 23.6 |
| pp62 (pp60) polyprotein precursor of p35 and p15 | CP530R | 60.5 |
| P12 attachment protein | O61R | 6.7 |
| P17. Required for progression of precursor membranes to icosahedral intermediates | D117L | 13.1 |
| J5R. Transmembrane domain | H108R | 12.5 |
| P54 (j13L). Binds to DLC1 chain (8 kd) of dynein, involved in virus entry. Required for recruitment of envelope precursors to the factory | E183L | 19.9 |
| J18L. Transmembrane domain | E199L | 22.0 |
| P14.5. DNA-binding. Required for movement of virions to plasma membrane | E120R | 13.6 |
| E248R (k2R). Possible component of redox pathway required disulfide bond formation. Structural protein | E248R | 27.5 |
| XP124L. Multigene family 110 member. Contains KDEL ER retrieval sequence and transmembrane domain | MGF 110-4L (XP124L) | 14.2 |
| EP402R. Similar to host CD2 protein. Required for binding red blood cells to infected cells and extracellular virus particles. Glycoprotein inserted into external virus envelope* | EP402R | 45.3 |
| Multigene family members | ||
| Multigene family 360 | MGF 360-1L (KP360L) | 41.7 |
| MGF 360-2L (KP362L) | 42.6 | |
| MGF 360-3L (L356L) | 41.7 | |
| MGF 360-4L (LIS382) | 44.9 | |
| MGF 360-5L (UP60L) | 7.0 | |
| MGF 360-6L (LIS375) | 43.9 | |
| MGF 360-7L (LIS375a) | 44.1 | |
| MGF 360-8L (J319L) | 31.3 | |
| MGF 360-9L (A125L) | 14.5 | |
| MGF 360-10L | 41.6 | |
| MGF 360-11L | 41.6 | |
| MGF 360-12L | 41.1 | |
| MGF 360-13L | 41.0 | |
| MGF 360-14L | 41.3 | |
| MGF 360-15R (A276R) | 31.6 | |
| MGF 360-16R (DP311R) | 35.6 | |
| MGF 360-17R (DP63R) | 8.4 | |
| MGF 360-18R (DP148R) | 17.2 | |
| MGF 360-19R (DP363R) | 42.4 | |
| MGF 360-20R (DP42R) | 4.9 | |
| MGF 360-21R | 42.0 | |
| MGF 360-22R | 41.7 | |
| Multigene family 110 | MGF 110-1L (L270L) | 32.4 |
| MGF 110-2L (U104L) | 12.2 | |
| MGF 110-3L (LIS124-1) | 14.3 | |
| MGF 110-4L (XP124L) | 14.2 | |
| MGF 110-5L (V82L) | 9.4 | |
| MGF 110-6L (Y118L) | 13.9 | |
| MGF 110-7L (LIS137) | 15.9 | |
| MGF 110-8L (LIS124-2) | 14.9 | |
| MGF 110-9L (LIS290) | 34.8 | |
| MGF 110-10L (190-2) | 22.7 | |
| MGF 110-11L (LIS119-1) | 32.5 | |
| MGF 110-12L (LIS 119-2) | 12.5 | |
| MGF 110-13L (LIS 117) | 18.3 | |
| MGF 110-14L (LIS121-2) | 14.7 | |
| Multigene family 300 | MGF 300-1L (J268L) | 31.3 |
| MGF 300-2R (J154R) | 17.6 | |
| MGF 300-3L (J104L) | 12.5 | |
| MGF 300-4L (J182L) | 21.7 | |
| Multigene family 505 | MGF 505-1R | 62.6 |
| MGF 505-2R (A489R) | 57.7 | |
| MGF 505-3R (A280R) | 32.5 | |
| MGF 505-4R (A505R) | 59.2 | |
| MGF 505-5R (A498R) | 58.7 | |
| MGF 505-6R (A518R) | 61.8 | |
| MGF 505-7R (A528R) | 61.7 | |
| MGF 505-8R | 61.7 | |
| MGF 505-9R (A506R) | 59.4 | |
| MGF 505-10R (A542R) | 59.4 | |
| MGF 505-11L (DP542L) | 63.1 | |
| Multigene family 100 | MGF 100-1R | 15.3 |
| MGF 100-2L (DP141L) | 16.8 | |
| MGF 100-3L (DP146L) | 17.2 |
* The table lists functions of ASFV-encoded proteins. Functions have been demonstrated experimentally for those proteins marked with an asterisk.
** Abbreviations: Herpes simplex virus (HSV); vaccinia virus (VV); methyl transferase (FTSJ); excision repair cross-complementation group (ERCC); yeast intermembrane mitochondrial protein (ERV).
Other predicted proteins encoded by the virus include enzymes involved in nucleotide metabolism (ribonucleotide reductase, thymidine kinase, thymidylate kinase and deoxyuridine triphosphatase), DNA replication and repair or transcription (DNA polymerase, DNA polymerase X, DNA ligase, topoisomerase II, guanylyl transferase, three members of DNA helicase superfamily II, and AP endonuclease). Deletion of the thymidine kinase, AP endonuclease and deoxyuridine triphosphatase genes does not affect virus replication in tissue culture cell lines but reduces virus replication in fully differentiated non-dividing macrophages and reduces virulence of the virus in pigs. Two enzymes involved in post-translational protein modification (a ubiquitin-conjugating enzyme and a serine/threonine protein kinase) and an enzyme involved in synthesis of isoprenoid compounds (trans-prenyltransferase), are encoded by the virus.
Five different multigene families (MGF 110, MGF 360, MGF 530/505, MGF 300 and MGF 100) are present in regions close to the genome termini. Large length variations between genomes of different isolates are due to gain or loss of members of these multigene families. MGF 110 contains 14 members, and individual isolates contain 5–11 of these. MGF 360 has 22 members, and 11–18 copies are encoded by different isolates. MGF 530/505 has 11 members, and 8–10 copies are present. MGF 300 has 4 members with 3–4 copies and MGF 100 has 3 members with 2–3 copies present. Members of families MGF 360 and 530 have been implicated as macrophage host range determinants, and deletion of 6 members of MGF 360 and 2 of MGF 530 results in an increase in type I interferon production (Afonso et al., 2004). These 6 copies of MGF 360 and 1–2 copies of MGF 530 are deleted from the genome of an attenuated field isolate OURT88/3 and the tissue-culture adapted BA71V isolate (Chapman et al., 2008).
Virus-encoded proteins that modulate the host response to infection include homologs of the apoptosis inhibitors Bcl-2 and IAP. Both of these proteins inhibit apoptosis; the IAP homolog inhibits caspase 3 activity. Viral Bcl-2 binds to most cellular proapoptotic proteins of the Bcl-2 family to inhibit apoptosis, and it also binds Beclin1 to inhibit autophagy (Galindo et al., 2008, Hernáez et al., 2013). A C-type lectin (pEP153R) has also been reported to inhibit apoptosis. Although the virus encodes proteins that inhibit apoptosis, apoptotic cells are nevertheless observed at late stages of infection. The A238L protein inhibits transcriptional activation dependent on a number of different host transcription factors, including NFAT, NFkB and cJun, by inhibiting their transactivation mediated by p300 (Granja et al., 2008). This protein also binds to and inhibits the host calcineurin phosphatase and is therefore predicted to inhibit calcineurin-dependent pathways (Miskin et al., 1998). The A238L protein may therefore inhibit transcriptional activation in infected macrophages of a wide range of host immunomodulatory genes that are dependent on these factors. A virus protein, EP402R, which is similar to the host T cell adhesion protein CD2, is required for the hemadsorption of red blood cells around virus-infected cells and is also thought to mediate the adhesion of extracellular virions to red blood cells. Deletion of the EP402R gene reduces virus dissemination in infected pigs and in vitro abrogates the ability of ASFV-infected cells to inhibit proliferation of bystander lymphocytes in response to mitogens. ASFV infection triggers ER stress and unfolded protein response (UPR). This leads to caspase 12 activation upregulation of the chaperones calnexin and calreticulin and the transcription factor 6 (ATF6) signaling pathway of the UPR but no other arms of the UPR (Galindo et al., 2012). One protein (designated pNL-S, pl14L or pDP71L) is similar over a conserved C-terminal domain to a herpes simplex virus-encoded neurovirulence factor (ICP34.5) and host protein GADD34 and is involved in ATF4 activation. These proteins all act to recruit cellular protein phosphatase 1 to dephosphorylate translation initiation factor eIF2α and inhibit global shut-off of translation and also thus inhibit CHOP induced apoptosis (Rivera et al., 2007, Zhang et al., 2010). GADD34 and ICP34.5 are larger and have other demonstrated roles. One of the major ASFV-induced proteins, encoded by gene CP204L, interacts with the heterogeneous nuclear ribonucleoprotein K (hnRNP-K), with potential implications in the downregulation of host cell mRNA translation. DP250R/g5R, which contains a NUDIX hydrolase domain, has been demonstrated to function as an ASFV decapping enzyme during infection, localizing together with both cellular and viral mRNAs and cap structure (Parrish et al., 2009, Quintas et al., 2017).
Lipids
Enveloped virions contain lipids, including glycolipids and phospholipids such as phosphatidylcholine, phosphatidylethanolamine and phosphatidylinositol.
Carbohydrates
One virion protein is known to be glycosylated (pEP402R), and glycolipids are also incorporated into virions. The virus encodes several predicted transmembrane proteins that contain putative N-linked glycosylation sites.
Genome organization and replication
The genomes of different ASFV isolates vary between 165,795 and 191,036 bp, excluding the terminal inverted repeat sequences, and encode 151–167 genes. (Figure 2 Asfarviridae). These ORFs are closely spaced with intergenic distances generally less than 200 nt, and read from both DNA strands. A few intergenic regions contain short tandem repeat arrays.
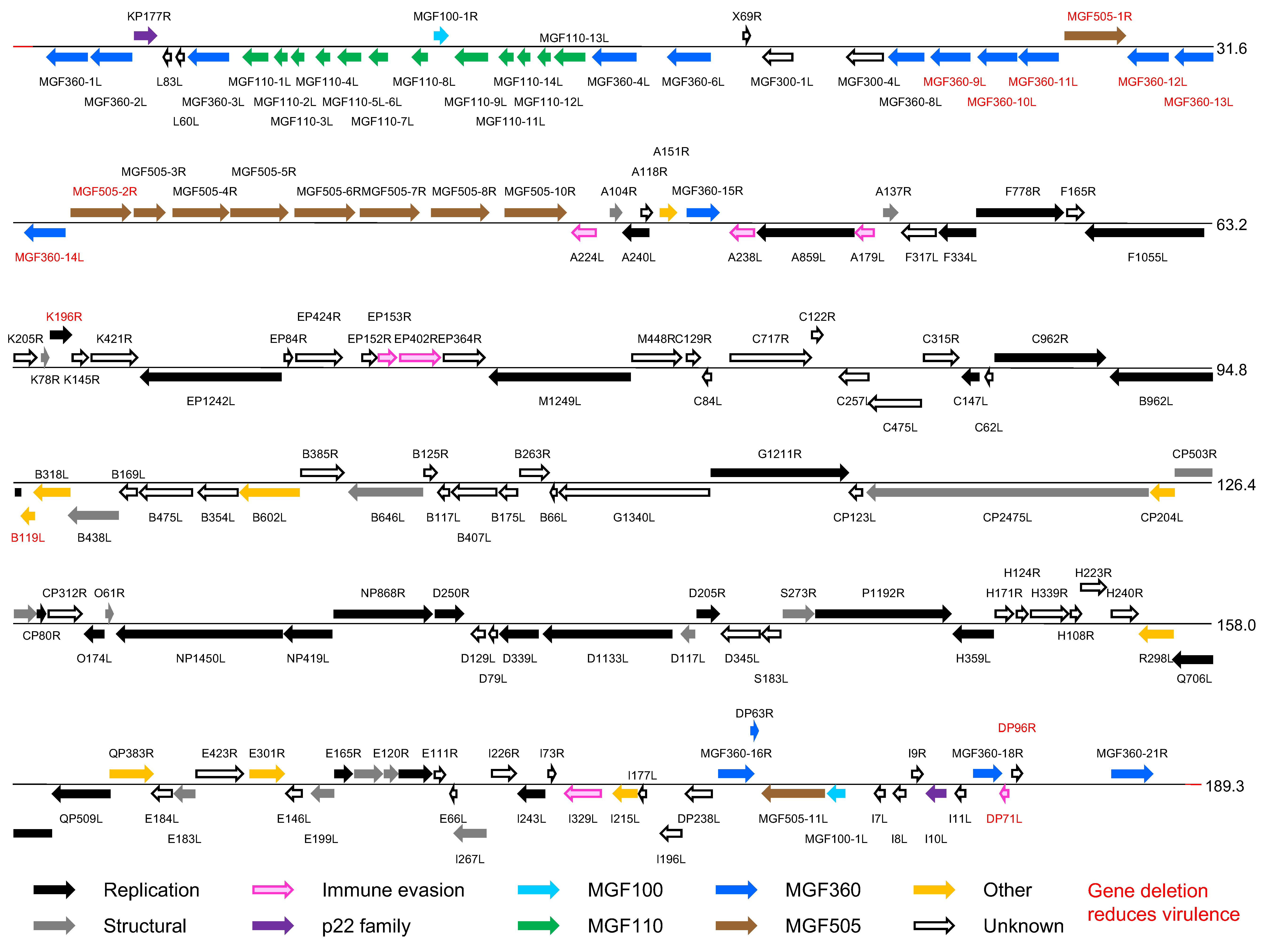 |
| Figure 2 Asfarviridae. Genomic organization of African swine fever virus isolate Georgia 2007/1. Open reading frames (ORFs) are shown by coloured arrows. They are closely spaced and present on either DNA strand, with short promoter sequences located close upstream. Transcription terminates at a sequence that consists of at least 7 T residues and that may be located some distance downstream from an ORF, resulting in mRNAs that read through more than one ORF. The 5′-ORF in such mRNAs is translated, but probably none downstream. mRNAs are modified by a 5′-cap structure and 3′-polyadenylation, and, since transcription takes place in the cytoplasm, are not spliced. Transcription and processing of mRNAs utilize virus-encoded enzymes and are independent of host RNA polymerase. (Adapted with permission from (Dixon et al., 2013)). |
The primary cell types infected by the virus include those of the mononuclear-phagocytic system, including fixed tissue macrophages and specific lineages of reticular cells. Virus replicates in vitro in macrophages and endothelial cells, and several isolates have been adapted to replicate in tissue culture cell lines. Virus enters cells primarily by clathrin- and dynamin-dependent receptor-mediated endocytosis, either in cooperation with the constitutive macropinocytosis of the macrophages or as an alternative entry mechanism (Figure 3. Asfarviridae). ASFV uncoating relies on host factors found at its endosomal passage and capsid disassembly occurs at the acid pH of the endosomal lumen (Cuesta-Geijo et al., 2012). Virion internal membrane fusion with endosomal membrane depends on viral protein pE248R (Hernáez et al., 2016) and on the integrity of cholesterol flux (Cuesta-Geijo et al., 2015). After capsid degradation and membrane fusion, viral cores exit endosomes. The ubiquitin-proteasome system is involved in the final degradation of the viral cores to set free the viral DNA in order to start replication (Barrado-Gil et al., 2017). Early mRNA synthesis begins immediately following entry, using enzymes and factors packaged in the virus core. Virus DNA replication and assembly take place in perinuclear factory areas, known as viral factories, where the virus is transported associated to the microtubular motor light chain dynein. At early times post-infection, virus DNA is detected in the nucleus, suggesting a possible role for nuclear enzymes in initial stages of DNA replication. Head-to-head virus DNA concatemers, which are thought to be replicative intermediates, are detected in the cytoplasm from 6 h post-infection. The mechanism of DNA replication in the cytoplasm is similar to that of viruses in the family Poxviridae.
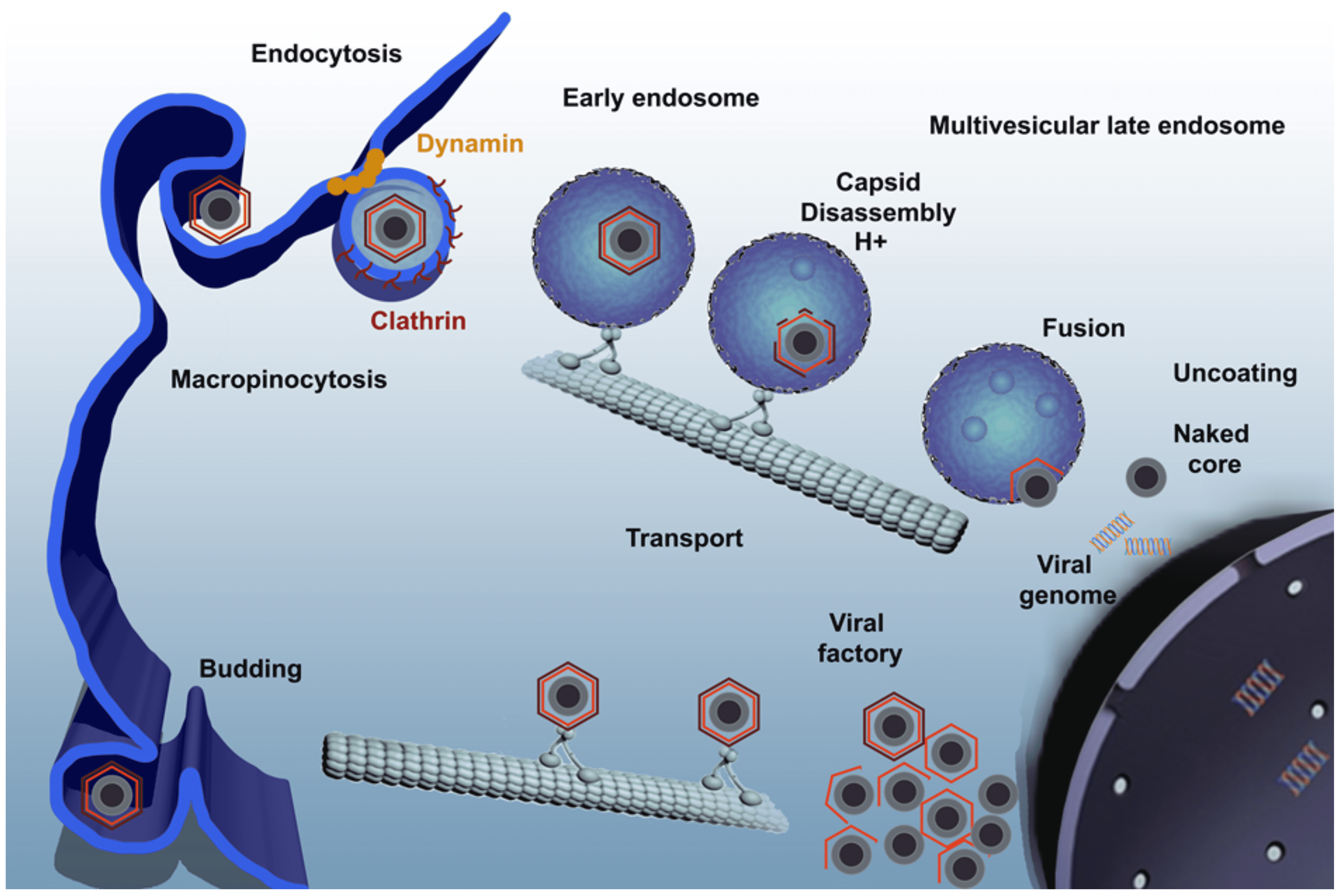 |
| Figure 3 Asfarviridae. African swine fever virus infectious cycle. Virus enters the cell by a complex process involving clathrin-mediated endocytosis and macropinocytosis. Virions progress rapidly through the endocytic pathway, undergoing uncoating. They reach mature endosomal compartments, where viral decapsidation and then fusion of the internal membrane with the endosomal membrane occur. This liberates viral cores to start replication. Replication and assembly takes place in a cytoplasmic site at the microtubule-organizing centre, giving rise to a structure known as a viral factory. Newly synthesized virions are assembled in the viral factory and exit the cell by budding. (Reprinted with permission from (Galindo and Alonso 2017)). |
Virus transcripts are 3′-polyadenylated and 5′-capped. Genes are expressed in an ordered cascade. Early genes are expressed prior to DNA replication; expression of late genes is dependent on the onset of DNA replication. Synthesis of some early genes continues throughout infection. Intermediate genes are expressed late but their expression does not depend on the onset of DNA replication. Promoter elements are relatively short and located immediately upstream from ORFs; transcription start sites are generally a short distance from start codons. Both early and late gene transcripts are of defined length; sequences of seven or more consecutive thymidylate residues in the coding strand are signals for mRNA 3′-end formation.
Several structural proteins are expressed as polyproteins and cleaved at the sequence GlyGlyX. The pp220 polyprotein precursor is myristylated. Other virus-encoded proteins are modified by phosphorylation (p10 and p32) and N-linked glycosylation. Virus morphogenesis takes place in perinuclear virus factories. Virus factories are surrounded by endosomes and membranes recruited at early times after infection, and intermediate filaments and mitochondria are recruited to them. Cholesterol and other lipids are required at different levels in the viral replication cycle (Cuesta-Geijo et al., 2015). A single lipid membrane apparently derived from the endoplasmic reticulum is incorporated as an internal lipid membrane in virus particles (Hawes et al., 2008). The P17 protein is essential for the progression of viral membrane precursors toward icosahedral intermediates. The P54 protein (pE183L) is required for intracellular virus transport and for recruiting envelope precursors to assembly sites. This protein binds to the DLC1 component of 8 kDa of the dynein motor complex, and this interaction is involved in recruitment to the assembly sites (Alonso et al., 2001). Disruption of this high affinity protein-protein interaction inhibits viral replication (Hernaez et al., 2010). Formation of the icosahedral capsid occurs on the internal membrane. Assembly of the major capsid protein P72 (pB646L) requires a virus-encoded chaperone pB602L. The pB438L protein is required for formation of the vertices of the icosahedral capsid. The virus genome and enzymes required to initiate infection are packaged into a nucleoprotein core (Salas and Andrés 2013). Processing of the virus polyproteins pp62 and pp220 is essential for core development (Suarez et al., 2010). Extracellular virus has a loose-fitting external lipid envelope derived by budding through the plasma membrane. Virus is transported to and from sites of assembly on microtubules. The pE120R virion protein is required for binding kinesin for virus transport on microtubules from assembly sites to the plasma membrane (Jouvenet et al., 2004).
Biology
ASFV infects domestic and wild swine (Sus scrofa domesticus and S. s. ferus), warthogs (Phacochoerus africanus) and bush pigs (Potamochoerus porcus). Disease signs are apparent only in domestic and wild swine. Soft ticks of the genus Ornithodoros are also infected, with O. moubata acting as a vector in parts of Africa south of the Sahara and O. erraticus having acted acting as a vector in south-west Spain and Portugal. Virus can be transmitted in ticks trans-stadially, and sexual and transovarial transmission has also been demonstrated in O. moubata. Warthogs, bush pigs and swine can be infected by bites from infected ticks. Virus is present in all excretions and secretions from infected pigs coincident with viremia. Transmission between domestic swine can occur by direct contact, by ingestion of infected meat, by fomites or mechanically by ticks (Olesen et al., 2017). Warthogs, bush pigs, wild swine and ticks act as reservoirs of virus. Disease has been endemic in many African countries and in Europe in Sardinia. In 2007, African swine fever was introduced to Georgia in the Trans-Caucasus and from there spread to neighbouring countries, including the Russian Federation and several neighboring countries in the eastern EU, such as the Baltic Republics, Poland, Czech Republic, Moldova and Romania. Disease was first introduced into Europe in Portugal in 1957 and was endemic in parts of the Iberian peninsula from 1960 until 1995. Sporadic outbreaks have occurred in, and been eradicated from, Belgium, Brazil, Cuba, the Dominican Republic, France, Haiti, The Netherlands and Malta, but otherwise have remained confined to Africa or the island of Sardinia.
ASFV causes haemorrhagic fever in domestic pigs and wild boar. Virus isolates differ in virulence and may produce a variety of disease signs ranging from acute to chronic to unapparent infection. Virulent isolates may cause 100% mortality in 5–15 days. Moderately virulent isolates have reduced mortality, and up to 50% of pigs may recover from infection. Chronic disease presents as characteristic arthritis, enlarged lymph nodes and renal dysfunction. Attenuated isolates cause may few disease signs. Recovered pigs can remain persistently infected and be protected against challenge with related virulent isolates. CD8+ T cells are required for this protection. Viruses replicate in cells of the mononuclear phagocytic system and reticuloendothelial cells in lymphoid tissues and organs of domestic swine. Cell surface markers expressed from intermediate stages of monocyte–macrophage differentiation are indicators of cell susceptibility to infection. Widespread cell death caused by apoptosis occurs in both T and B lymphocytes in lymphoid tissues and endothelial cells in arterioles and capillaries. This accounts for the lesions seen in acute disease. Disseminated intravascular coagulation develops during the late phase of acute infections, and this may lead to the characteristic haemorrhagic syndrome.
Vaccine approaches
Conventional approaches for vaccine development, such as inactivated vaccines, have failed against ASFV, independently of the inactivation method and the adjuvant used (Blome et al., 2014). Conversely, live attenuated viruses (LAV) have been used as experimental models to induce efficient protection against homologous ASFV challenges. Most of the current knowledge about the mechanisms involved in ASFV protection is due to the use of LAVs. There is no doubt that both antibodies and CD8+ cells play a role in protection (Escribano et al., 2013, Onisk et al., 1994, Oura et al., 2005), together with the induction of an efficient innate immune response. Classical attenuated AFV strains, either isolated from the field in endemic situations or obtained through serial passage in tissue culture, opened the door to more sophisticated LAVs obtained by specific gene targeting using recombination technologies. Thus, recombinant LAVs obtained by disruption of multiple MGF members or the 9GL ORF from Georgia07 strain, which is currently circulating in Eastern Europe, have rendered good protection levels against homologous challenge (O'Donnell et al., 2015). Interestingly, serial deletion of two of these genes, 9GL and UK (DP96R), rendered safer efficient vaccine prototypes than those lacking the single ORFs (O'Donnell et al., 2017). Also, manipulation of other attenuated and virulent strains has generated LAV prototypes conferring protection (Reis et al., 2017, Sánchez-Cordón et al., 2017). BA71∆CD2, a LAV obtained by deleting the hemagglutinin gene from the BA71 virulent strain is capable of conferring protection against the homologous virus and also against other genotype I strains and genotype II virus Georgia07 (Monteagudo et al., 2017). Despite their safety, subunit vaccines are still one step behind LAVs. Several antigens have been proved to contribute to partial protection, including the structural P54, P72, P30 proteins and hemagglutinin. These antigens elicited both neutralizing antibodies (Gómez-Puertas et al., 1998, Ruíz-Gonzalvo et al., 1996) and, in some cases, CD8+ T cells (Argilaguet et al., 2012, Lacasta et al., 2014). More research is needed to develop effective subunit vaccines, and in the long term these may provide a better option than LAVs (Arias et al., 2017).
Antigenicity
Antibodies induced in pigs that recover from infection with less virulent ASFV isolates can neutralize virus infection in pig macrophages and cell lines (Escribano et al., 2013). The differences in lipid composition among wild-type and cell culture adapted virions correlates with susceptibility to virus neutralization by antibodies (Gómez-Puertas et al., 1997). Serotyping of virus isolates by neutralization has not been yet accomplished, and there is no evidence to relate the 23 genotypes described based on the sequence of the gene encoding the capsid protein P72 (Bastos et al., 2003) with serotypes. Some virus protein targets for neutralizing antibodies have been identified using antibodies against proteins P72, P30 and P54 (Gómez-Puertas et al., 1998). The neutralization mediated by protein P30 inhibits virus internalization rather than attachment. Protein P12 has also been described as an additional virus attachment protein, despite the fact that neutralizing antibodies against this protein have not been yet demonstrated. Recently, the ASFV CD2v and C-type lectin gene loci have been identified as possible mediators of serological specificity.
Derivation of names
Asfar: from African swine fever and related viruses.
Relationships within the family
There is a limited extent of variability observed within the family (Figure 4 Asfarviridae).
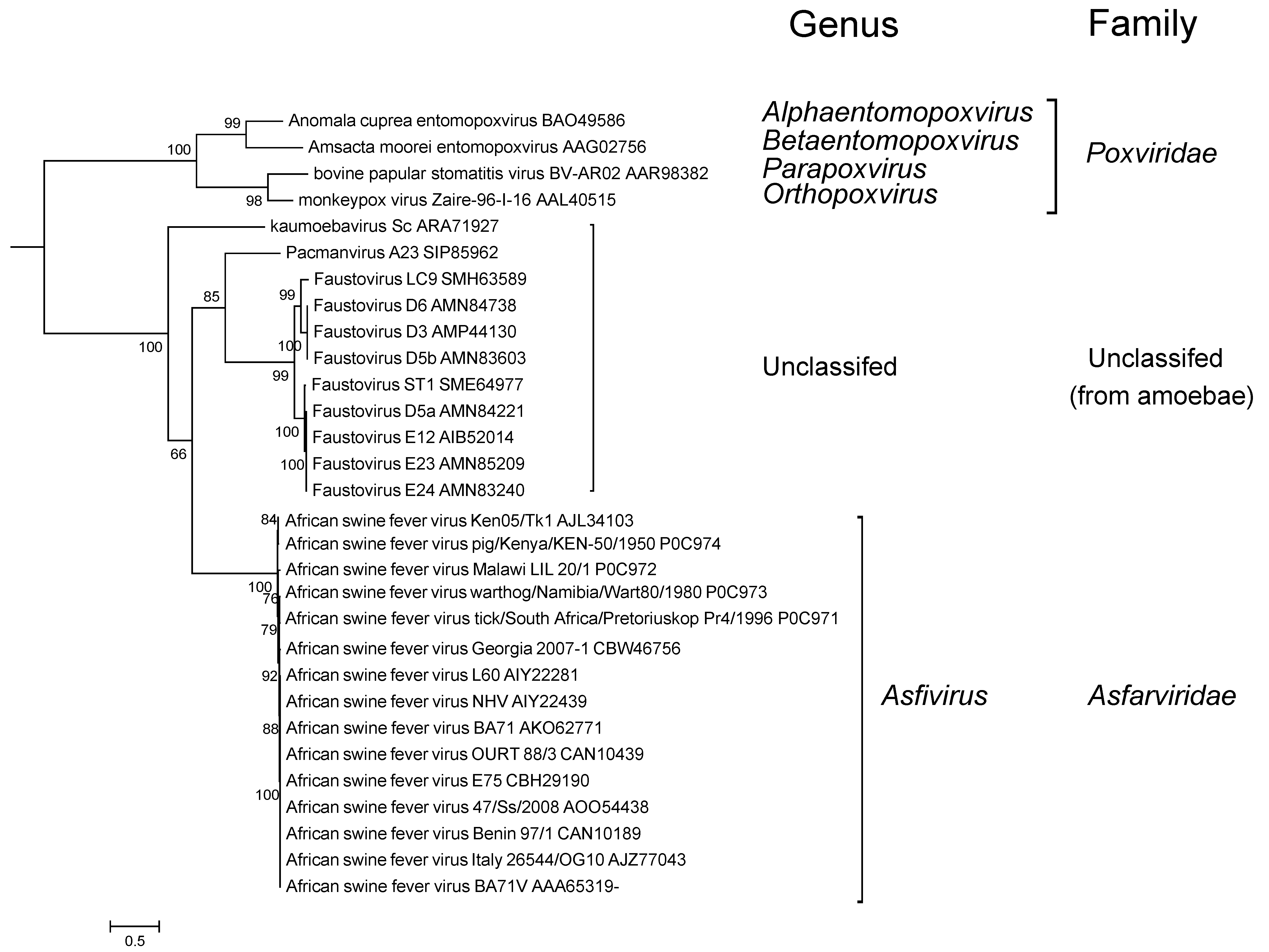 |
| Figure 4 Asfarviridae. Phylogenetic analysis of African swine fever virus and related viruses. A maximum likelihood tree was produced based on DNA polymerase B protein sequences aligned using MUSCLE in the MEGA6.0 package (Tamura et al., 2013). The tree was visualized using iTolV3. Branches supported by bootstrap values higher than 50% are indicated (kindly provided by J. Andreani, Aix-Marseille Université, France). This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
ASFV was listed as a member of the family Iridoviridae until 1984, when it was removed as a member of the unassigned genus African swine fever virus group, (renamed African swine fever-like viruses in 1995). The genus was renamed Asfivirus and placed in the new family Asfarviridae in 1998. Analysis of replication strategies and virus genes have shown that ASFV is related to other viruses in the nucleo-cytoplasmic large DNA virus “superfamily”, which includes the families Poxviridae, Iridoviridae, Phycodnaviridae and Mimiviridae.
Metagenomic sequences related to ASFV have been identified in virus fractions from oceans, sewage and human serum (Loh et al., 2009). ASFV-like sequences (FJ957903-38) were detected in a metagenomic study of freshwater ponds in the Mississippi delta (Wan et al., 2013). A total of 48 of the sequences obtained showed similarity to 23 ASFV genes. However, the amino acid sequences imputed from these 48 matches were highly divergent from ASFV sequences.
Faustovirus, kaumoebavirus and Pacmanvirus are large viruses that infect amoeba, and are distantly related to ASFV (Figure 4. Asfarviridae) with about 30 related genes (Andreani et al., 2017). However, their genomes are considerably larger (about 400 kbp) than that of ASFV (170-194 kbp), and they are not known to replicate in vertebrates.
Faustovirus (CZDJ02000001) has a double-stranded DNA genome of 466,000 bp with 451 predicted ORFs sharing distant similarity with ASFV (Klose et al., 2016). A 17,000 bp region includes five putative ORFs that are related to parts of the ASFV major capsid protein. However, the architecture of the related gene of the prototypic Faustovirus E12 (Reteno et al., 2015) is clearly different from that of ASFV as it contains putative introns and exons. In contrast, capsid-encoding genes with introns and exons can be found in other viruses of amoeba such as a mimivirus infecting Acanthamoeba polyphaga. Faustovirus virions have an icosahedral shape and a unique double layered protein capsid. The presence of an inner and outer capsid (with double jelly-roll folds) with spikes and the absence of an internal membrane are other distinct features of the Faustovirus virion. Faustovirus infects amoeba (Vermamoeba vermiformis) (Reteno et al., 2015) and was detected in biting midges (Culicoides imicola) (Temmam et al., 2015).
Pacmanvirus (LT706986) has a linear genome of at least 395,000 bp with 465 genes, of which 31 are similar to those of Faustovirus and kaumoebavirus. The major capsid protein locus of Pacmanvirus appears to be different from those of kaumoebavirus and Faustovirus. It infects Acanthamoeba castellanii (Andreani et al., 2017).
Kaumoebavirus (the name derived from King Abdulaziz University amoeba virus) has 250 nm icosahedral capsids and a DNA genome of at least 350,000 bp including 465 genes (KX552040) (Bajrai et al., 2016). The closest matches to kaumoebavirus proteins are for proteins encoded by Faustovirus and African swine fever virus, although the genome organization is distinct. The kaumoebavirus genome contains introns and exons, similar to that of Faustovirus.