Family: Benyviridae
David Gilmer and Claudio Ratti
The citation for this ICTV Report chapter is the summary published as Gilmer et al., (2017):
ICTV Virus Taxonomy Profile: Benyviridae, Journal of General Virology, 98: 1571–1572.
Corresponding author: David Gilmer ([email protected])
Edited by: Hélène Sanfaçon and Stuart G. Siddell
Posted: July 2017
PDF: ICTV_Benyviridae.pdf
Summary
The Benyviridae is a family of plant viruses with rod-shaped virions (Table 1. Benyviridae). Genomes are multipartite single stranded, positive sense RNAs with 5′ m7G cap and 3′ polyA and there is post-translational cleavage of the viral replicase. The better-known members are transmitted by root-infecting vectors in the family Plasmodiphorales, once described as fungi but now classified as Cercozoa. Beet necrotic yellow vein virus is the cause of ‘rhizomania’ disease of sugar beet. This widespread soil-borne disease can severely reduce sugar yields.
Table 1. Benyviridae. Characteristics of members of the family Benyviridae.
| Characteristic | Description |
| Typical member | beet necrotic yellow vein virus isolate Japan S (RNA1: D84410; RNA2: D84411; RNA3: D84412; RNA4: D84413; RNA5: D63936), species Benyvirus necrobetae |
| Virion | Non-enveloped, rod shaped particles about 20 nm in diameter and up to about 390 nm long with two or more modal lengths |
| Genome | At least two segments of polyadenylated positive-sense RNA (approximately 7 and 4.6 kb). Up to 3 additional RNA components of 1.3-1.8 kb each may also be present |
| Replication | Cytoplasmic, probably associated with the endoplasmic reticulum. |
| Translation | Directly from genomic or intracellular subgenomic RNAs |
| Host Range | Plants |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Kitrinoviricota, class Alsuviricetes, order Hepelivirales: 1 genus containing 4 species |
Virion
Morphology
Non-enveloped, helically constructed rod-shaped particles, with an axial canal and of up to five different lengths have been described (Figure 1. Benyviridae). In beet necrotic yellow vein virus (BNYVV), the predominant lengths are about 390, 265, 100, 85 and 65-80 nm and their diameter is about 20 nm. The right-handed helix with a pitch of 2.6 nm has an axial repeat of four turns, involving 49 CP subunits, each occupying four nucleotides (Steven et al., 1981). Coat proteins constitute about 95% of the particle weight.
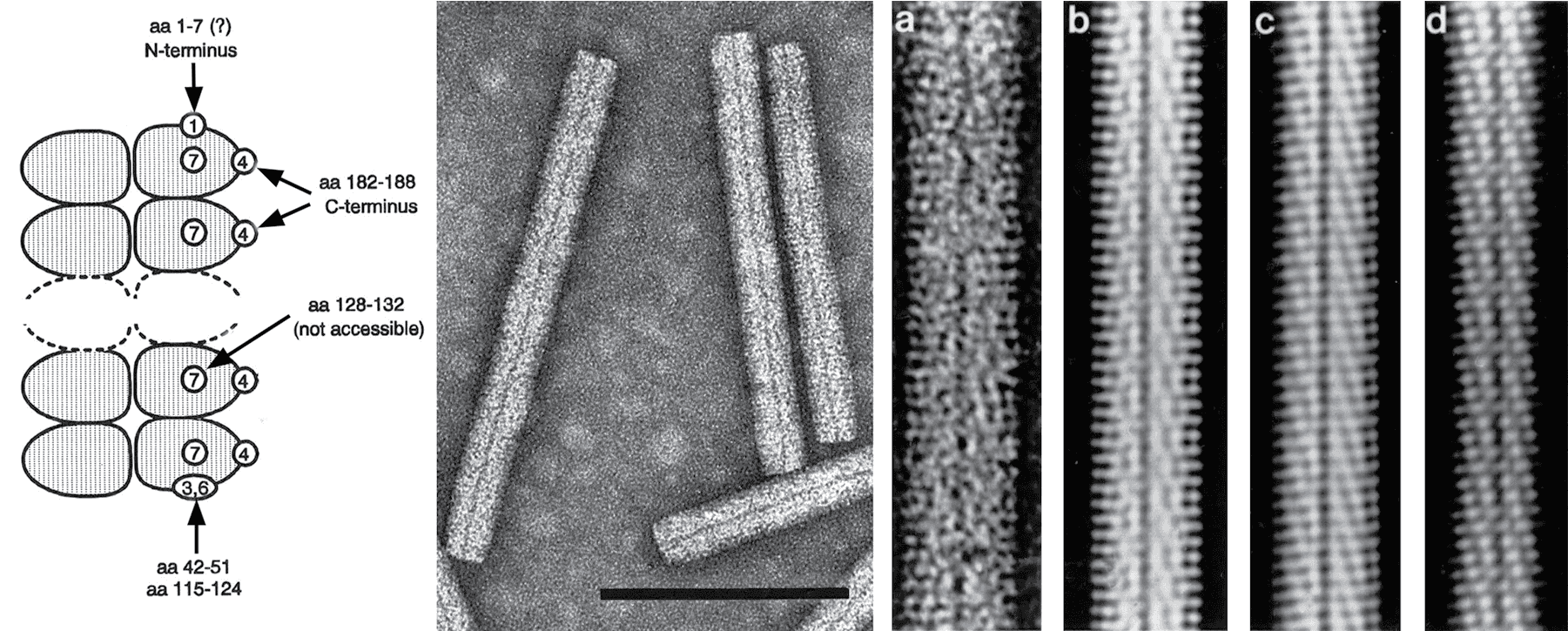 |
| Figure 1. Benyviridae. (Left) Scheme showing the accessibility to antibodies of various parts of the coat protein amino acid (aa) sequence in particles of BNYVV. Encircled numbers designate different epitopes. (Center) Negative contrast electron micrograph of stained purified particles of BNYVV. (Right) From left (a) negative contrast electron micrograph of a BNYVV particle and (b, c, d) computer-filtered micrographs of BNYVV particles (modified from (Steven et al., 1981), courtesy of A.C. Steven). The bar represents 100 nm. |
Physicochemical and physical properties
The viral particles are sometimes unstable in sap with strong infectivity losses within one to five days at room temperature.
Nucleic acid
BNYVV has four or five linear positive sense ssRNAs of about 6.7, 4.6, 1.8, 1.4 and 1.3 kb, respectively (Kiguchi et al., 1996, Saito et al., 1996). The viral RNAs are capped at the 5′ end and, unlike the RNAs of all other rod-shaped plant viruses, are 3′-polyadenylated. Viral RNAs have a conserved 3′ structure essential for RNA replication initiation. After mechanical transmission to laboratory test plants, BNYVV RNA3, RNA4 and RNA5 may carry deletions or may be lost entirely (Bouzoubaa et al., 1991). Isolates of beet soil-borne mosaic virus (BSBMV) have four RNAs (Heidel et al., 1997) while rice stripe necrosis virus (RSNV) and burdock mottle virus (BdMV) apparently only contain two genomic RNAs (Lozano and Morales 2009, Kondo et al., 2013).
Proteins
The major CP species is 21–23 kDa in size. The minor CP amber readthrough protein (c. 75 kDa) is detected near one extremity of BNYVV particles and initiates virus assembly. The minor capsid protein contains a KTER motif in its C terminal part that is necessary for the transmission of the virions by Polymyxa betae (Tamada et al., 1996).
Genome organization and replication
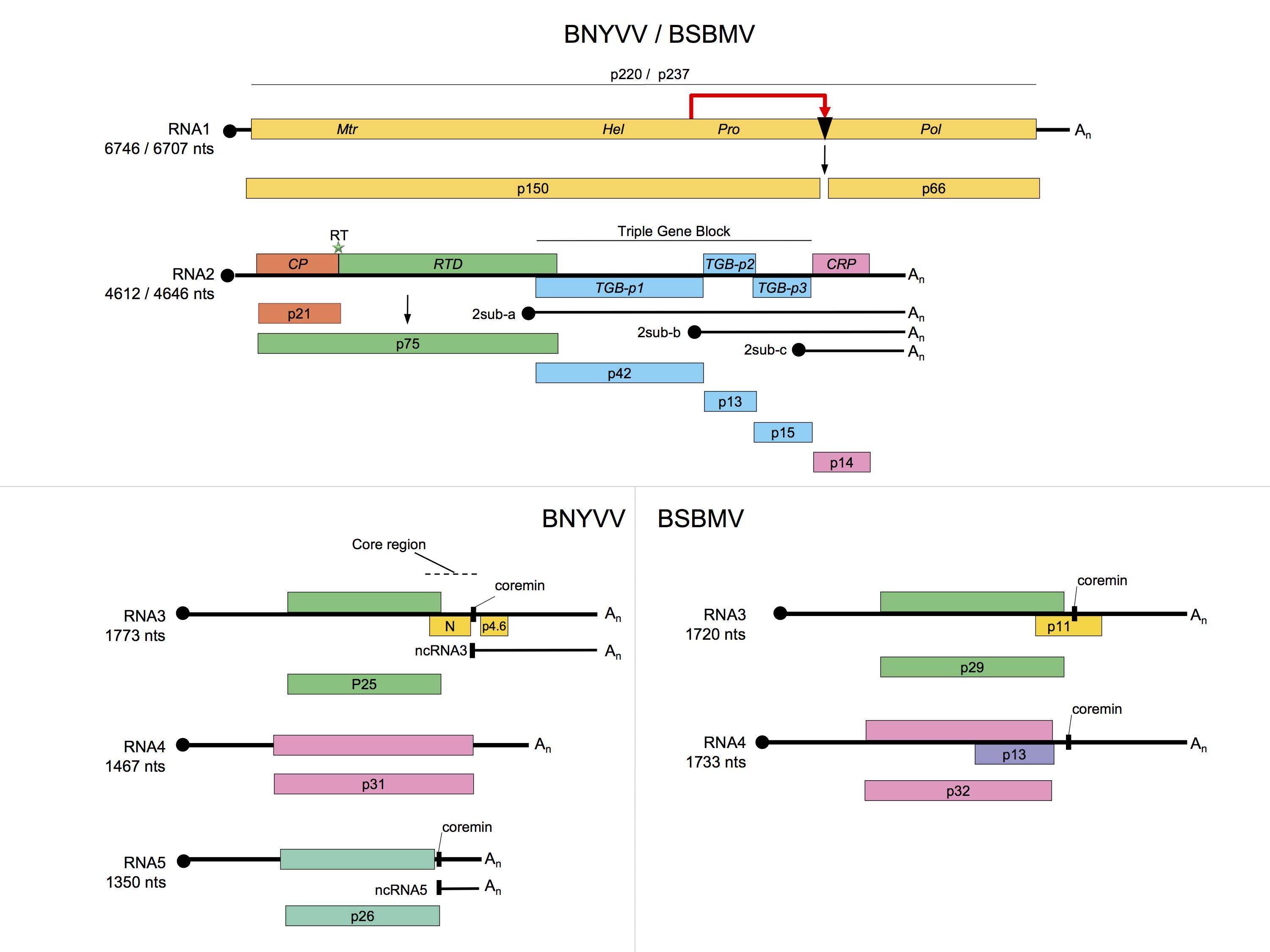
RNA1 contains one large ORF coding for a replication-associated protein that is cleaved post-translationally. This proteolytic cleavage of the replicase distinguishes the benyviruses from all other viruses with rod-shaped particles, which have their replication-associated proteins encoded in two ORFs. The replication complex is located to the ER of infected cells by sequences in both the N-terminal region and the RdRp domain of the polyprotein (Pakdel et al., 2015). In vitro translation of BNYVV RNA1 may initiate at two sites: at the first AUG in the sequence, at position 154, or at a downstream AUG at position 496. The resulting proteins of 237 and 220 kDa, respectively, both contain methyltransferase (Mtr) motifs in their N-terminal part, helicase (Hel) and papain-like protease motifs (Prot) in their central part, and RNA dependent RNA polymerase (RdRp) motifs in their C-terminal part (Figure 2. Benyviridae). RNA2 of BNYVV contains six ORFs; namely, the CP gene which is terminated by a suppressible amber stop codon (UAG), the CP readthrough protein gene, the TGB coding for TGB1, TGB2 and TGB3 (42, 13 and 15 kDa respectively) and a gene coding for the P14 cysteine-rich protein, a viral post-transcriptional gene-silencing suppressor (14 kDa). The TGB ORFs and the P14 gene are expressed by means of subgenomic RNAs (Figure 2. Benyviridae)(Chiba et al., 2013). The three triple gene block (TGB)-encoded proteins (TGB1, TGB2 and TGB3) but not the CP are necessary for cell-to-cell movement (Gilmer et al., 1992) while long distance movement requires CP. BNYVV TGB1 labeled with green fluorescent protein (GFP) on its N-terminus is targeted by TGB2 and TGB3 to punctate bodies associated with plasmodesmata. The N-terminal region of the BNYVV TGB1 has nucleic acid binding activity and its C-terminal region contains consensus sequence motifs characteristic of an ATP/GTP-dependent helicase. The three proteins are found along membrane-rich peripheral bodies, thought to be derived from endoplasmic reticulum (ER). The movement protein (MP) of tobacco mosaic virus can functionally complement TGB proteins. RNA1 and RNA2 are sufficient for replication of BNYVV in the local lesion host Chenopodium quinoa or in the systemic hosts Spinacia olacera and Nicotiana benthamiana. The typical rhizomania symptoms in beet are produced only in the presence of RNA3; RNA4 greatly increases the transmission rate by P. betae and RNA5 may modulate the type of symptoms that result. RNA3 and RNA4 are always present in natural BNYVV infections. BNYVV is able to replicate and encapsidate BSBMV RNA3 and RNA4 (Ratti et al., 2009, D'Alonzo et al., 2012).
Biology
BNYVV has been found at the cytoplasmic surface of mitochondria soon after infection. Later, virions of most BNYVV isolates are scattered throughout the cytoplasm of infected cells or occur in aggregates. Masses of particles arranged in parallel or angle-layer arrays may also be formed. Depending on the isolate, only one or both types of aggregates occur. Membranous accumulations of ER may also be found. The p25 protein encoded in RNA3, and in particular the sequence variability of a tetrad (four amino acids, residues 67-70), appears to be involved in the resistance breaking capabilities of the virus on sugar beet.
The natural host ranges of benyviruses are very narrow. Beta vulgaris is the natural host of BNYVV and BSBMV. Species of Chenopodium are infected experimentally, often only locally.
RNA3 is essential for the viral long distance movement in Beta sp. thanks to the accumulation of a noncoding RNA3, ncRNA3, (Lauber et al., 1998, Peltier et al., 2012) produced by 5’-3’ exoribonuclease cellular activity that is stalled by a structural motif involving the “coremin” motif. Nicotiana benthamiana, Spinacia olacera and Beta macrocarpa are systemic hosts for BNYVV and BSBMV. In nature, BNYVV and BSBMV are transmitted by P. betae and rice stripe necrosis virus (RSNV) by P. graminis. The viruses are also mechanically transmissible.
BNYVV has spread to most sugar beet-growing areas worldwide. Variants (A-, B- or P-type) occur in different geographical areas and are distinguished by minor changes in the CP sequence, by the tetrad sequence in the p25 ORF and by the presence of the RNA5 species. BSBMV is widely distributed in the United States. RSNV occurs in Africa, and South and Central America. Burdock mottle virus (BdMV) has been identified in a restricted area in Japan.
Antigenicity
BNYVV and BSBMV are serologically distinct. Epitope mapping has revealed portions of the BNYVV CP that are either exposed along the entire particle length, e.g. the immunodominant C-terminus, or are accessible only on one extremity or after disruption of the particles (Figure 1. Benyviridae).
Derivation of names
Beny: from beet necrotic yellow vein virus.
Relationships within the family
There is <60% amino acid identity between the CP amino acid sequences of BNYVV and BSBMV; other members of the genus are more distantly related (Figure 3. Benyviridae). The percentages of identity between various non-structural proteins of the two viruses range between 38% (cysteine-rich protein) and 84% (replication-associated protein). RNA species of BNYVV and BSBMV share common structural motifs at their extremities but the coding sequences differ. BNYVV p26 protein resembles BSBMV p29 (Figure 2. Benyviridae) in sequence and functions. The “coremin” sequence of 20 nucleotides involved in the systemic movement of the virus is present on BNYVV RNA3 core sequence and RNA5, BSBMV RNA3 and RNA4 (Figure 2. Benyviridae) as well as on cucumber mosaic virus and tomato aspermy virus (Bromoviridae) RNAs. The CP of BdMV shares 38% sequence identity with that of BNYVV. TGB1 of BdMV shares approximately 50% amino acid sequence identity with the corresponding proteins of BNYVV and BSBMV while RSNV RNA1 shares about 52% sequence identity with the RNA1s of BNYVV and BSBMV.
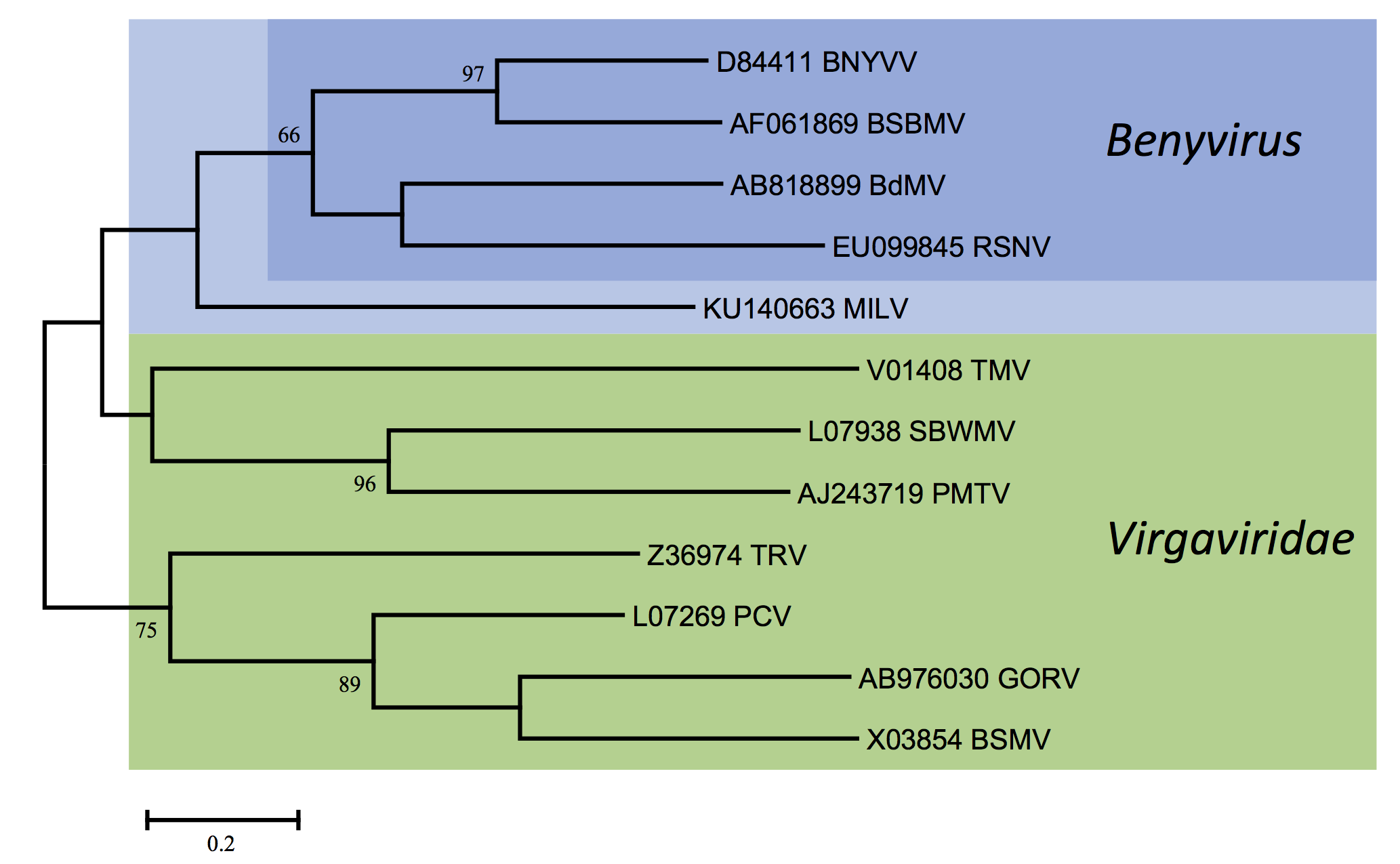 |
| Figure 3. Benyviridae. Phylogenetic tree based on the codon-aligned nucleotide sequences of the capsid proteins of benyviruses and other rod-shaped plant viruses. The analyses were done in MEGA7 using the Maximum Likelihood method based on the Tamura-Nei model and 1000 bootstrap replicates. The tree with the highest log likelihood (-6317.4271) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches (where >60%). The scale indicates the number of substitutions per site. All positions containing gaps and missing data were eliminated. Viruses (or possible members) of the genus Benyvirus are highlighted (BdMV: burdock mottle virus; BNYVV: beet necrotic yellow vein virus; BSBMV: beet soil-borne mosaic virus; MILV: Mangifera indica latent virus; RSNV: rice stripe necrosis virus). Abbreviations of the other viruses (all family Virgaviridae) are: BSMV: barley stripe mosaic virus (Hordeivirus); GORV: gentian ovary ringspot virus (Goravirus); PCV: peanut clump virus (Pecluvirus); PMTV: potato mop top virus (Pomovirus); SBWMV: soil-borne wheat mosaic virus (Furovirus); TMV: tobacco mosaic virus (Tobamovirus); TRV: tobacco rattle virus (Tobravirus). This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Benyviruses are morphologically similar to other rod-shaped viruses that are classified in the family Virgaviridae (furo-, gora-, peclu-, pomo-, hordei-, tobra- and tobamoviruses). However, the Mtr, Hel and RdRp motifs in the replication-associated proteins show a higher degree of similarity to those of some viruses of the families Togaviridae (rubella virus) and Hepeviridae (human hepatitis E virus) than to those of other rod-shaped plant viruses (Figure 4. Benyviridae). The coat proteins of the rod-shaped viruses are distantly related (Figure 3. Benyviridae) and contain conserved residues, e.g. RF and FE in their central and C-terminal regions, respectively, which are presumably involved in the formation of salt bridges. Like pomo-, peclu- and hordeiviruses, but unlike furo-, tobamo- and tobraviruses, the benyviruses have their movement function encoded on a triple gene block. Sequence identities in the first and second triple gene block-encoded proteins reveal affinities not only to pomo- and hordeiviruses, but also, more distantly, to viruses in the families Alphaflexiviridae and Betaflexiviridae, including potex- and carla-viruses (Figure 5. Benyviridae).
 |
| Figure 4. Benyviridae. Phylogenetic tree based on the codon-aligned nucleotide sequences of the RdRp protein domains of benyviruses and other related viruses. The analysis and abbreviations are as in Figure 3. Benyviridae and the tree with the highest log likelihood (-21239.3562) is shown. Additional virus abbreviations are: HBBLV: Hubei beny-like virus 1 (unclassified); HEV: human hepatitis E virus (Orthohepevirus, Hepeviridae); MPTLV: Macrophomina phaseolina tobamo-like virus (unclassified); RUBV: rubella virus (Rubivirus, Togaviridae). This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
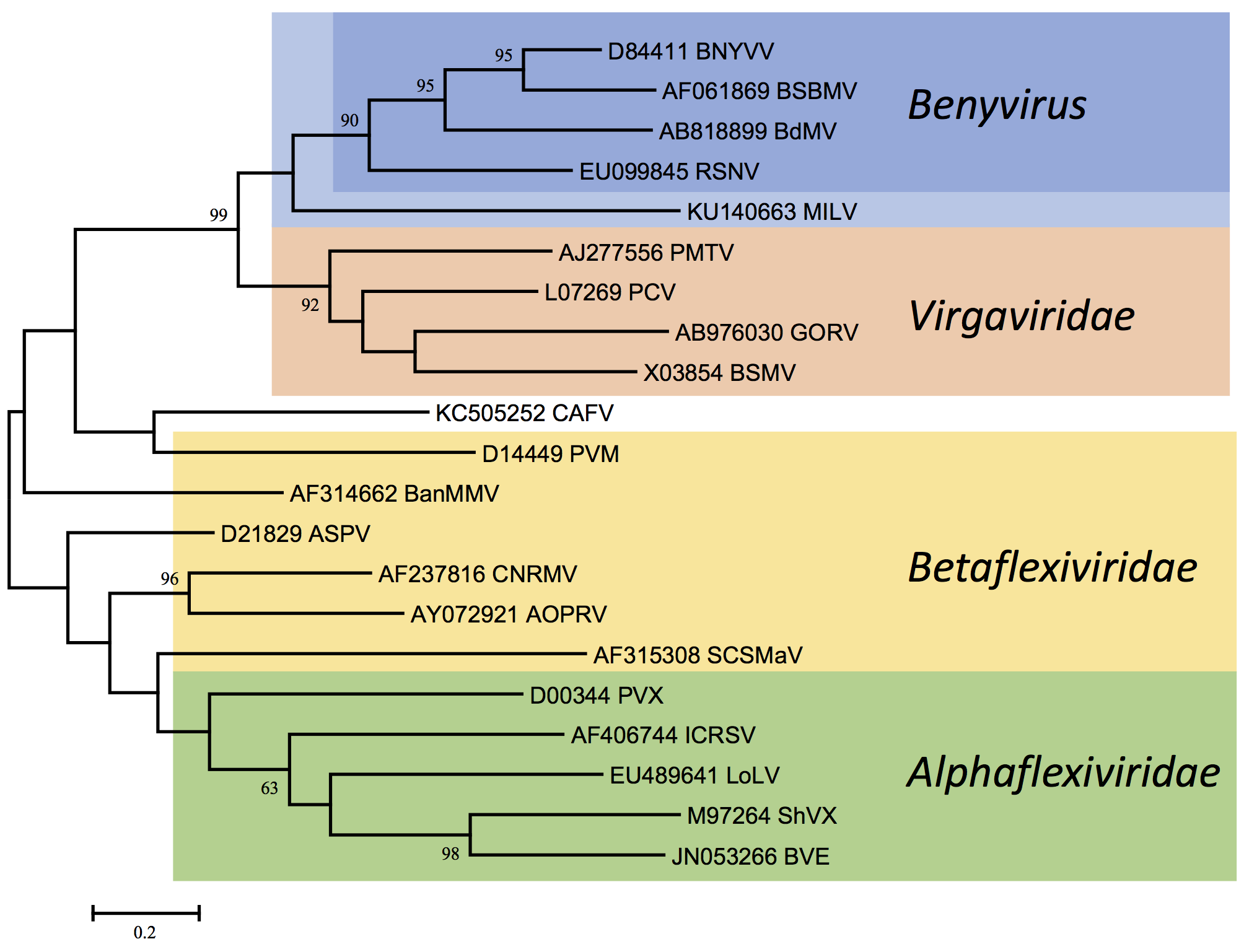 |
| Figure 5. Benyviridae. Phylogenetic tree based on the codon-aligned nucleotide sequences of the first triple-gene block proteins of benyviruses and other plant viruses. The analysis and abbreviations are as in Figures 3. Benyviridae and 4. Benyviridae and the tree with the highest log likelihood (-13812.5960) is shown. Additional virus abbreviations are: AOPRV: African oil palm ringspot virus, Robigovirus; ASPV: apple stem pitting virus, Foveavirus; BanMMV: banana mild mosaic virus, unassigned Betaflexiviridae; BVE: blackberry virus E, unassigned Alphaflexiviridae; CAFV: cassava alphaflexivirus, unassigned Alphaflexiviridae; CNRMV: cherry necrotic rusty mottle virus, Robigovirus; ICRSV: Indian citrus ringspot virus, Mandarivirus; LoLV: Lolium latent virus, Lolavirus; PVM: potato virus M, Carlavirus; PVX: potato virus X, Potexvirus; SCSMaV: sugarcane striate mosaic-associated virus, unassigned Betaflexiviridae; ShVX: shallot virus X, Allexivirus. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |