Family: Clavaviridae
David Prangishvili, Tomohiro Mochizuki, Ying Liu and Mart Krupovic
The citation for this ICTV Report chapter is the summary published as Prangishvili et al., (2019):
ICTV Virus Taxonomy Profile: Clavaviridae, Journal of General Virology, 100, 1267–1268.
Corresponding authors: David Prangishvili ([email protected]) and Mart Krupovic ([email protected])
Edited by: Andrew M. Kropinski and Stuart G. Siddell
Posted: June 2019
PDF: ICTV_Clavaviridae.pdf
Summary
The family Clavaviridae includes viruses that replicate in hyperthermophilic archaea from the genus Aeropyrum (Table 1 Clavaviridae). The non-enveloped rigid virions are rod-shaped, about 143×16 nm, with terminal cap structures, one of which is pointed and carries short fibers, while the other one is rounded. The virion displays helical symmetry and is constructed from a single major α-helical protein, which is heavily glycosylated, and several minor capsid proteins. The 5,278 bp, circular, double-stranded DNA genome of Aeropyrum pernix bacilliform virus 1 is packed inside the virion as a left-handed superhelix and held in place by interactions with the molecules of the major capsid protein.
Table 1 Clavaviridae. Characteristics of members of the family Clavaviridae
| Characteristic | Description |
| Typical member | Aeropyrum pernix bacilliform virus 1 (AB537968), species Clavavirus yamagawaense |
| Virion | Bacilliform particles, 143 nm long and 16 nm in diameter, with one end pointed and the other one rounded |
| Genome | Circular double-stranded DNA of 5,278 bp |
| Replication | Non-lytic, chronic infection |
| Translation | Not characterized |
| Host range | Hyperthermophilic archaea from the genus Aeropyrum |
| Taxonomy | Single genus with a single species |
Virion
Morphology
Virions of Aeropyrum pernix bacilliform virus 1 (APBV1) are non-enveloped, rigid rod-shaped particles, about 143 x 16 nm, terminating with cap structures (Table 1 Clavaviridae, Figure 1 Clavaviridae) (Mochizuki et al., 2010, Ptchelkine et al., 2017). The cylindrical virion body has a helical C5 symmetry, whereas the caps display five-fold symmetry (Ptchelkine et al., 2017). One of the caps is pointed and carries five short fibers with globular protruding domains, whereas the other cap is rounded and devoid of visible fibers (Figure 1. Clavaviridae). The cap structures likely play a role in DNA packaging and host recognition. The virion architecture and manner of DNA packaging are unprecedented among viruses of bacteria and eukaryotes (King et al., 2012, Prangishvili et al., 2017).
 |
| Figure 1 Clavaviridae. Electron micrograph of virions of Aeropyrum pernix bacilliform virus 1 (APBV1) and three-dimensional reconstruction of the virion structure. (a) Virions imbedded in vitreous ice (b) 3D model of the virion shown from the side; scale bars, 10 nm. Top (c) and side (d) view of APBV1 map (transparent surface) and model of the major capsid protein VP1. VP1 subunits related by C5 symmetry are colored in red, yellow, green, orange, and magenta. Reproduced with permission from (Ptchelkine et al., 2017). |
Physicochemical and physical properties
Virions are highly thermostable and remain infectious after incubation at 100°C for three hours; however, purified cell-free particles lose activity after overnight incubation at 90°C or autoclaving at 120°C for 20 minutes (Mochizuki et al., 2010).
Nucleic acid
Virions contain a single molecule of circular double-stranded DNA of 5,278 bp. The G+C content of the genome is 52.7%, which is somewhat lower than that for the host chromosome (Mochizuki et al., 2010).
Lipids
Not detected.
Proteins
The virions carry multiple copies of a single major capsid protein VP1 of about 10 kDa and three minor capsid proteins which have molecular masses in the range of 9.5–21.5 kDa (Mochizuki et al., 2010). The minor capsid proteins, VP2–4, are believed to form the terminal caps of the virion. VP1 (81 aa), encoded by ORF6-81, consists of two α-helices linked with a beta hairpin, a structural fold not seen in the capsid proteins of other known viruses (Ptchelkine et al., 2017). Each VP1 molecule makes extensive hydrophobic contacts to six other neighbouring subunits (Figure 2. Clavaviridae), forming an impeller blade-like arrangement in the virion (Ptchelkine et al., 2017). Positively-charged residues of VP1 interact with the phosphodiester backbone of the double-stranded DNA genome and facilitate its packaging as a left-handed superhelix (Ptchelkine et al., 2017). The three minor capsid proteins appear to be paralogous to the major capsid protein (Ptchelkine et al., 2017); thus, all genes encoding structural proteins might have evolved through gene duplication.
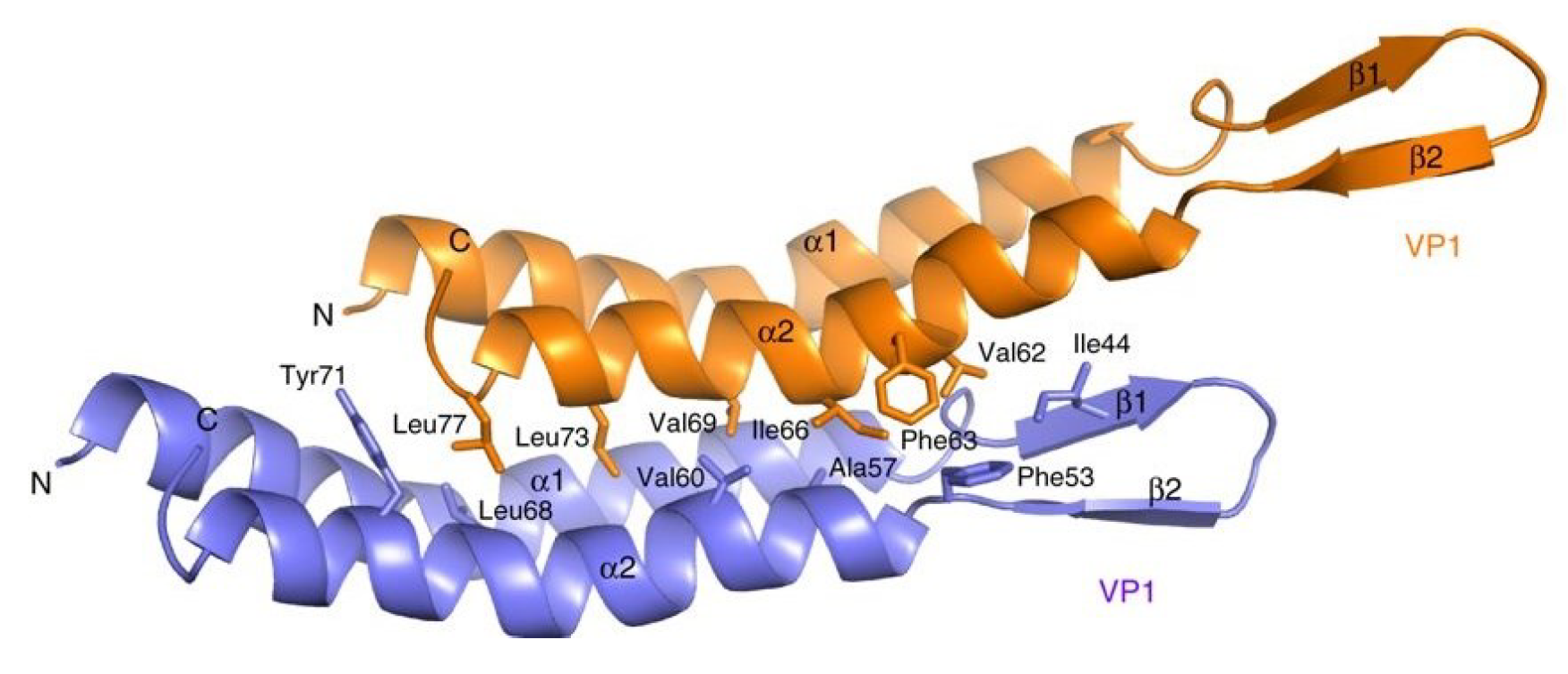 |
| Figure 2 Clavaviridae. The structure of the Aeropyrum pernix bacilliform virus 1 major capsid protein VP1. Two neighboring VP1 molecules are shown in orange and blue, respectively, and residues involved in intermolecular contacts are labeled. These residues create multiple hydrophobic contacts and a large interface. Reproduced with permission from (Ptchelkine et al., 2017). |
Carbohydrates
The major capsid protein is heavily glycosylated, whereas VP2, VP3 and VP4 do not appear to be post-translationally modified (Mochizuki et al., 2010).
Genome organization and replication
The genome contains 14 open reading frames (ORFs) larger than 40 codons, all of which are located on the same DNA strand (Figure 3 Clavaviridae) (Mochizuki et al., 2010). None of the putative gene products share similarity with sequences in public databases (Krupovic et al., 2018). Genes encoding four structural proteins have been determined by protein sequencing.
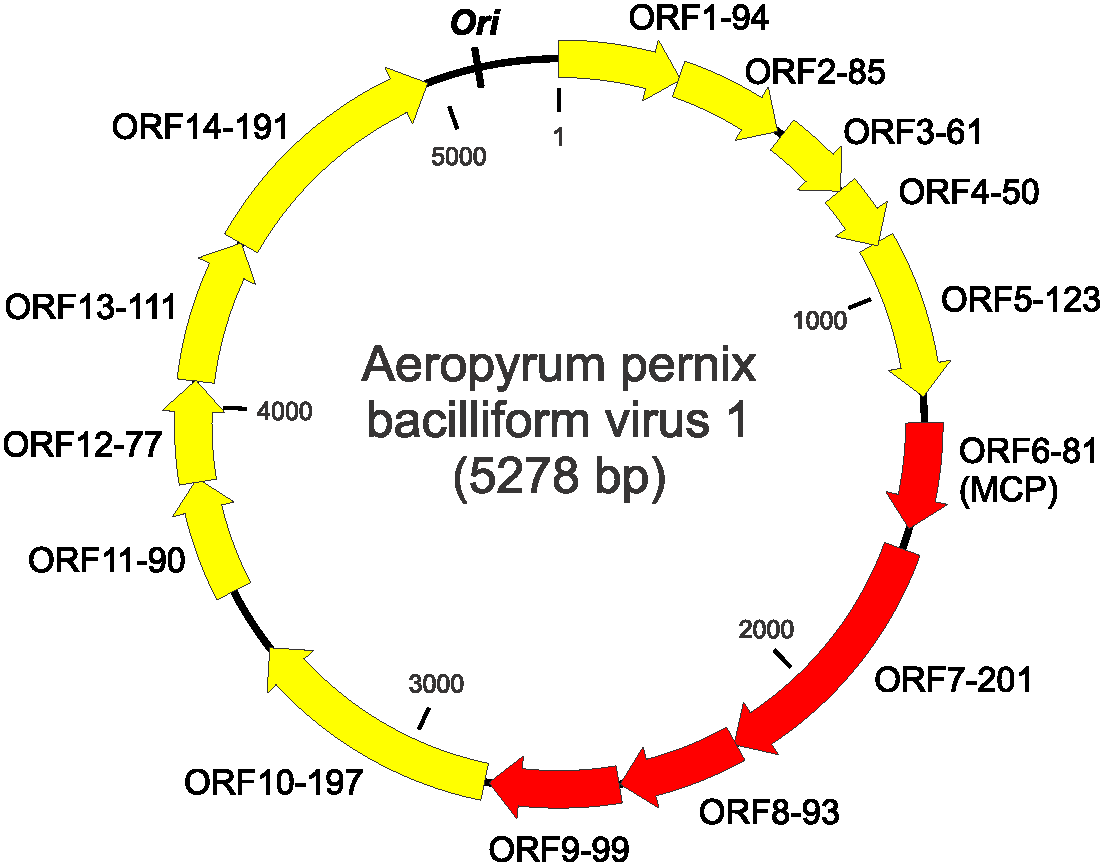 |
| Figure 3 Clavaviridae. Genome map of Aeropyrum pernix bacilliform virus 1. Each ORF is designated with two numbers; the first number is the order starting from the first ORF after the putative ori, and the second reflects the number of amino acids in the encoded proteins. Red arrows indicate genes encoding virion proteins. MCP- major capsid protein. |
The origin of genome replication , ori, is most likely located in the intergenic region between ORF1-94 and ORF14-191 (Figure 3. .Clavaviridae). Moreover, this region contains a perfect inverted repeat of 19 nucleotides (5′-TGTAGTACACACAATAATT-N68-AATTATTGTGTGTACTACA-3′) (Mochizuki et al., 2010), but the function of the repeat remains unclear. APBV1 does not encode identifiable DNA and RNA polymerases and is likely to depend on the host machineries for genome replication and transcription.
Biology
The virus was isolated from a coastal hot spring with temperature reaching 100°C at Yamagawa, island of Kyushu, Japan (Mochizuki et al., 2010). The virus host Aeropyrum pernix, a member of the hyperthermophilic archaeal order Desulfurococcales within the phylum Crenarchaeota, is the most thermophilic species among known obligative aerobic organisms, growing optimally at 90–95°C. APBV1 virions are released without apparent host cell lysis. The virus does not encode recognizable integrases; integration of the virus genome into the host chromosome has not been detected.
Derivation of names
Clava: from Latin clava, for “club”, “stick”.
Relationships within the family
APBV1 is the sole known member of the family.
Relationships with other taxa
The family Clavaviridae comprises a single genus Clavavirus, with one species, Clavavirus yamagawaense. No relationships have been revealed with other viruses based on structural or sequence similarities.

