Family: Closteroviridae
Marc Fuchs, Moshe Bar-Joseph, Thierry Candresse, Hans J. Maree, Giovanni P. Martelli, Michael J. Melzer, Wulf Menzel, Angelantonio Minafra and Sead Sabanadzovic
The citation for this ICTV Report chapter is the summary published as Fuchs et al., (2020):
ICTV Virus Taxonomy Profile: Closteroviridae, Journal of General Virology, 101: 364–365
Corresponding author: Marc Fuchs ([email protected])
Edited by: F. Murilo Zerbini and Sead Sabanadzovic
Posted: January 2020
PDF: ICTV_Closteroviridae.pdf
This work is dedicated to the memory of our friend and colleague Professor Giovanni Paolo Martelli, former chair of the Closteroviridae Study Group and life member of the ICTV, who passed away in January 2020.
Summary
Plant viruses in the family Closteroviridae possess long, helically constructed filamentous particles (650–2,200 nm in length) and a large positive-sense single-stranded, mono-, bi- or tripartite RNA genome (13,000 to nearly 19,000 nucleotides) (Table 1. Closteroviridae). The presence of a cellular HSP70 homolog and a duplicated, diverged copy of the capsid protein (minor capsid protein, CPm) genes in the virus genome are hallmarks of the family. The genome expression strategy is based on proteolytic processing, a +1 ribosomal frameshift, and sub-genomic messenger RNAs. Members of the family are classified into four genera: Ampelovirus, Closterovirus, Crinivirus and Velarivirus. Their genetic diversity is primarily influenced by strong negative selection and recombination. Viruses are mostly phloem-restricted and induce specific cytoplasmic aggregates of virus particles intermingled with membranous vesicles derived from the endoplasmic reticulum and possibly mitochondria. Transmission is by aphids, whiteflies, pseudococcid mealybugs or soft scale insects in a semi-persistent manner. Experimental transmission by mechanical inoculation is very difficult or impossible, and seed transmission is not known. A few members in the family have been engineered as gene expression and RNA interference vectors.
Table 1. Closteroviridae. Characteristics of members of the family Closteroviridae
| Characteristic | Description |
| Typical member | beet yellows virus (AF056575), species Closterovirus flavibetae |
| Virion | Non-enveloped, flexible filaments of 650–2,200 nm in length and 12 nm in diameter |
| Genome | 13–19.3 kb of positive-sense, mono-, bi- or tripartite RNA |
| Replication | In association with vesicular membranes derived from the endoplasmic reticulum and mitochondria |
| Translation | Proteolytic processing, a +1 ribosomal frameshift, and sub-genomic messenger RNAs |
| Host range | Plants (mainly dicots), transmitted by aphids, whiteflies, mealybugs and soft scales. No seed transmission |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Kitrinoviricota, class Alsuviricetes, order Martellivirales, family includes seven genera, and 58 species |
Virion
Morphology
Virions are helically constructed filaments with a pitch of the primary helix in the range of 3.4–3.8 nm, containing about 10 protein subunits per turn of the helix and showing a central hole of 3–4 nm (Figure 1. Closteroviridae, top). The very flexuous and open structure of the particles is the most conspicuous trait of members of the family (Agranovsky 2016). Virions have a diameter of about 12 nm and lengths ranging from 650 nm (viruses with a fragmented genome) to over 2,200 nm (viruses with a monopartite genome). The fragility of virions and a tendency to end-to-end aggregation contribute to the fact that many viruses are reported with a range of lengths. Two types of coat proteins (CP), the major CP and a CP analog or minor CP (CPm), are the most abundant protein components involved in the formation of (most) closterovirid particles. CPm encapsidates the 600–700 5′-terminal nucleotides of the virus RNA, at one extremity (75–100 nm) of the particle, as shown for beet yellows virus (BYV), carrot yellow leaf virus (CYLV), citrus tristeza virus (CTV), grapevine leafroll-associated virus 2 (GLRaV-2) and lettuce infectious yellows virus (LIYV), thus forming a distinct structure for which the terms “rattlesnake”, “heterodimeric” or “bipolar” have been coined (Figure 2. Closteroviridae) (Agranovsky 2016).
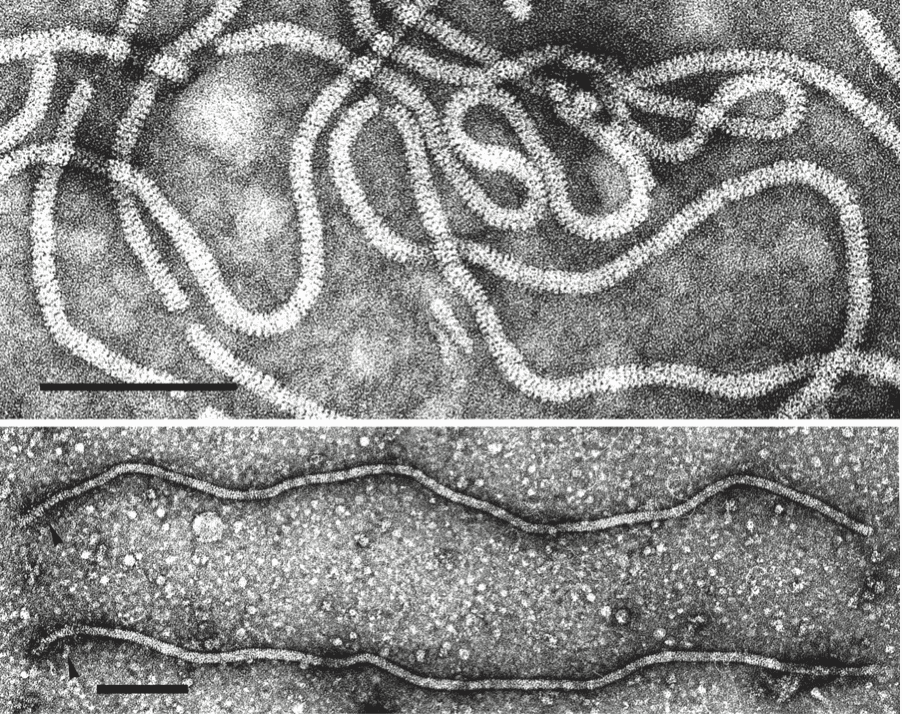 |
| Figure 1. Closteroviridae. (Top) Negative-contrast electron micrograph of virions of citrus tristeza virus (CTV) (genus Closterovirus). Particles of members of the genera Ampelovirus and Crinivirus have a similar morphology. The bar represents 100 nm (Courtesy of R.G. Milne). (Bottom) Beet yellows virus (BYV) particles showing a decorated extremity (arrow heads) following exposure to an antiserum to the N-terminal peptide expressed from the CPm gene. The bar represents 100 nm (Courtsey of D.E. Lesemann). |
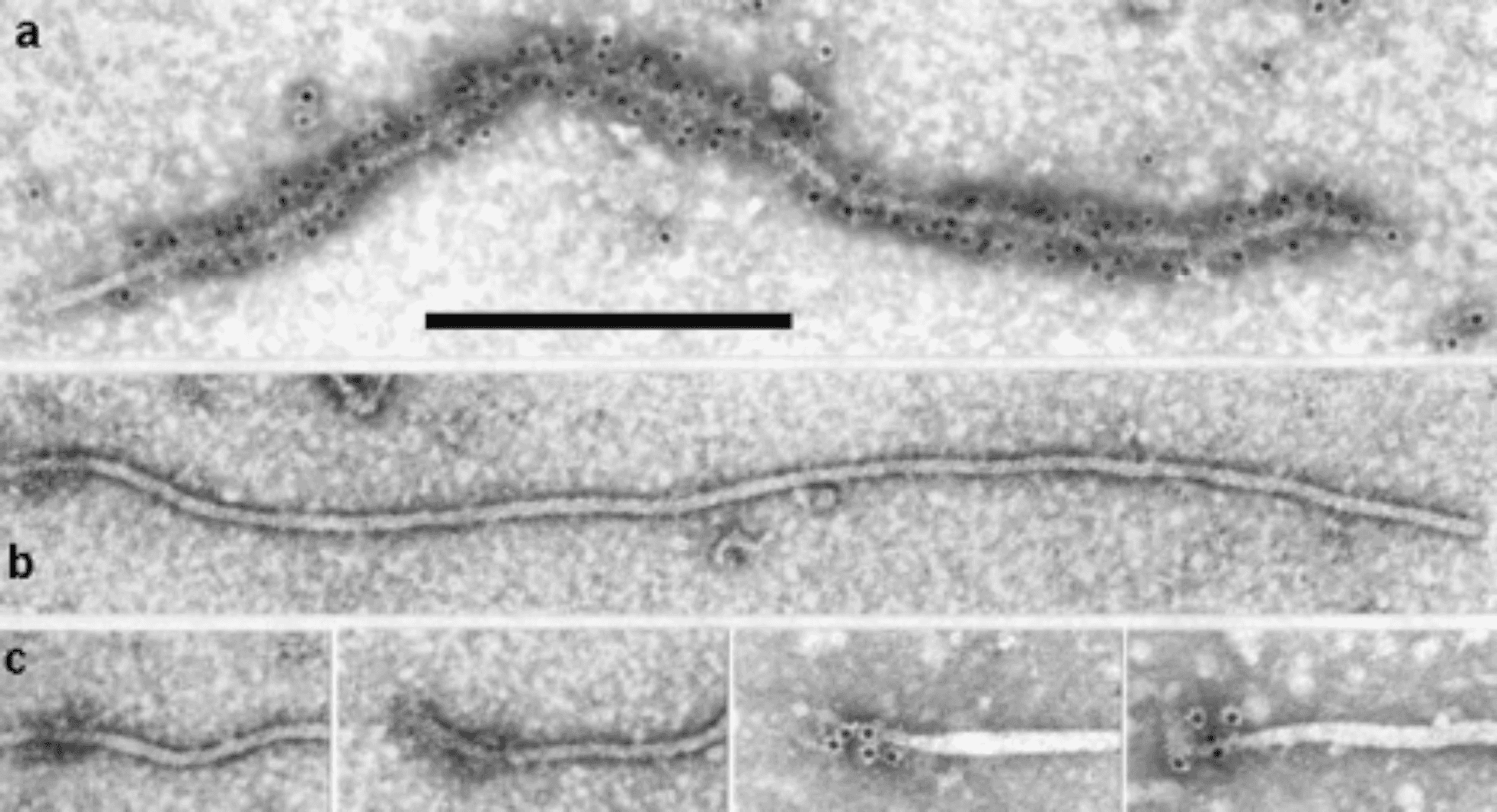 |
| Figure 2. Closteroviridae. Closterovirus virion structure and organization. Electron micrographs of virions of beet yellows virus (genus Closterovirus) negatively-stained and decorated with an antiserum specific to (a) coat protein (CP; note that the CPm tail is not decorated), and (b) CPm at the 75 nm tail of a virus particle. (c) Close-up of the 75 nm tail of four selected particles decorated with an antiserum specific to CPm. Scale bar represents 300 nm (from (Agranovsky and Lesemann 2000) with permission). |
Physicochemical and physical properties
Virions usually sediment as a single band in sucrose or Cs2SO4 gradients. S20,w is around 130–140, buoyant density is 1.33 g cm−3 in CsCl and 1.257 g cm−3 in Cs2SO4. Virions of several members are degraded by CsCl and are unstable in high salt concentration, resist moderately high temperatures (thermal inactivation is around 45–55 °C) and organic solvents, but are sensitive to RNase and chelation.
Nucleic acid
Regardless of whether the genome is monopartite or fragmented, virions contain a single molecule of linear, positive-sense, single-stranded RNA, constituting 5–6% of the particle weight. Genome size is related to particle length, ranging from 13,000 to slightly over 19,000 nucleotides. The 5′-end of the genome is likely capped. The 3′-end is not polyadenylated and does not possess a tRNA-like structure, but has several hairpin structures and a putative pseudoknot essential for replication (Agranovsky 2016).
Proteins
Structural proteins of most members of the family consist of a major CP and a diverged copy of it, denoted minor CP (CPm), with masses ranging from 22 to 46 kDa (CP) and 23 to 80 kDa (CPm), according to species. A group of ampeloviruses with a small genome (about 13,000 nt) apparently lack a true CPm. For BYV, and presumably for most other members of the family, CPm is required for the assembly of the 5′-extremity of the virion, and the ~60 kDa protein (p55-p64) is required for incorporation of both the 70 kDa heat shock protein homolog (HSP70h) and the CPm into virions, which also incorporate a 20 kDa protein (p20) that may form the tip segment of the virion head (Agranovsky 2016).
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
Members of the family have one of the largest genomes among plant viruses, reflecting duplication and the acquisition of nonviral coding sequences (e.g., protease, HSP70 protein) via RNA recombination (Figure 3. Closteroviridae). Recombination may also explain differences in genome organization between members of different genera and members of the same genus (Agranovsky 2016, Dolja et al., 2006, Karasev 2000, Rubio et al., 2013, Ruiz et al., 2018). The complex ORF1a-ORF1b invariably encodes the replication-related proteins, with methyl-transferase (Mtr), helicase (Hel) and RNA-directed RNA polymerase (RdRP) conserved domains. Downstream ORFs, which encode in a 5′ to 3′ direction a 6 kDa small hydrophobic protein, HSP70h, a ~60 kDa protein, CP and CPm, form a five-gene module which is conserved, with few modifications, among most members of the family (Agranovsky 2016, Dolja et al., 2006). The HSP70h and the ~60 kDa proteins are integral virion components present in all members of the family for which the particle structure was studied. The functions postulated for HSP70h include mediation of cell-to-cell movement through plasmodesmata, involvement in the assembly of multi-subunit complexes for genome replication and/or subgenomic RNAs synthesis, and assembly of virus particles (Agranovsky 2016, Dolja et al., 2006). The ~60 kDa protein is required for incorporation of both HSP70h and CPm into virion heads. The duplication of the capsid protein gene seems to be unique among viruses with elongated particles (Agranovsky 2016, Dolja et al., 2006). In general, capsid proteins and their homologs (CPm) show a significant degree of sequence conservation, and the duplicate copies probably retain the general spatial folding and some other crucial properties of the CPs. A notable exception is a group of ampeloviruses with the smallest genomes in the family [e.g. grapevine leafroll-associated virus 4 (GLRaV-4) and related viruses, pineapple mealybug wilt-associated virus 1 (PMWaV-1) and pineapple mealybug wilt-associated virus 3 (PMWaV-3)] which do not appear to possess CPm. For lettuce infectious yellows virus (LIYV), the replication of both genomic RNAs is asynchronous as genomic RNA1 and sub-genomic RNAs (sgRNAs) accumulate before significant accumulation of genomic RNA2 can be detected, suggesting that RNA1 likely replicates in cis while RNA2 replicates in trans. Knockout mutation studies showed that the single-stranded RNA-binding protein p34 that is encoded by RNA1 is a trans enhancer of RNA2 replication (Kiss et al., 2013).
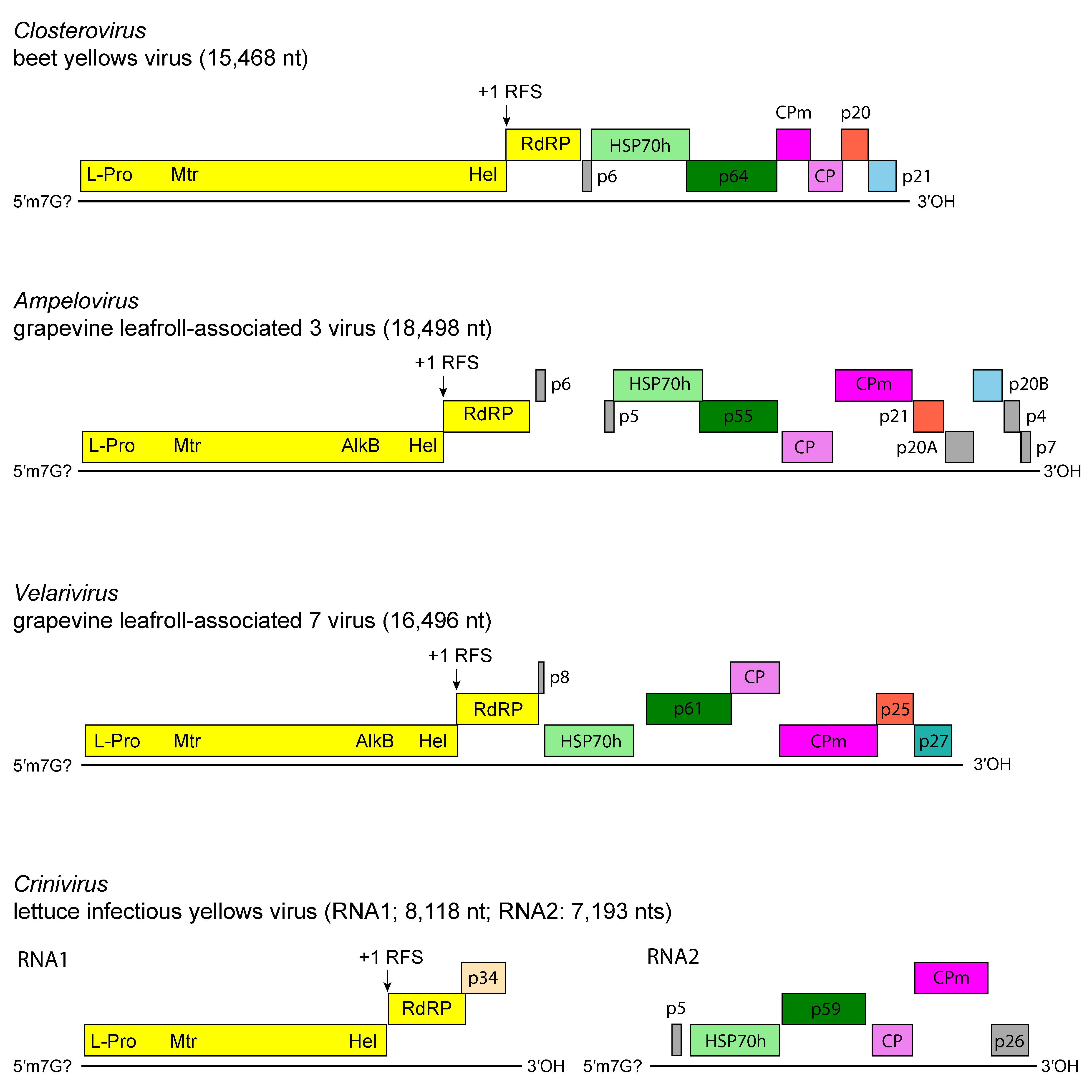 |
| Figure 3. Closteroviridae. Genome organization of representative members of the family Closteroviridae. L-Pro, leader proteinase; Mtr, methyltransferase; Hel, helicase; RdRP, RNA-directed RNA polymerase; HSP70h, heat shock protein 70 homolog; ~60 kDa protein; CP, coat protein; CPm, minor coat protein. |
The genome expression strategy is based on: (i) proteolytic processing of the polyprotein encoded by ORF1a; (ii) +1 ribosomal frameshift for the expression of the RdRP domain encoded by ORF1b, a mechanism not found in other positive-sense RNA plant viruses; and (iii) expression of the downstream ORFs via the formation of a nested set of 3′-co-terminal sgRNAs) (Agranovsky 2016, Dolja et al., 2006, Karasev 2000, Rubio et al., 2013). The double-stranded (ds) RNA patterns are very complex and variable among species, reflecting the different numbers and sizes of the ORFs present in individual genomes and, in some cases, the existence of defective RNAs (D-RNAs). Replication occurs in the cytoplasm, possibly in association with endoplasmic reticulum-derived membranous vesicles and vesiculated mitochondria. From an evolutionary point of view, closteroviruses represent a monophyletic virus lineage that might have evolved from a smaller filamentous virus when higher plants differentiated (Agranovsky 2016, Dolja et al., 2006). This progenitor virus, thought to be composed of three genes encoding replication-associated proteins, a protein (p6) with affinity for cell membranes, and a single CP, acquired the HSP70h and a ~60 kDa protein derived from a fusion of two domains, an N-terminal domain of unknown evolutionary provenance, and a duplicated CP domain. Under the pressure of further modular evolutionary events, i.e. duplication of the CP gene, acquisition of diverse suppressors of RNA silencing and of additional genes acquired via horizontal gene transfer (e.g. papain-like cysteine proteinase and AlkB domains), this family ancestor gave rise to the progenitors of the four extant genera of the family. Viruses belonging to one of these genera (Crinivirus), differentiated further by the genome splitting into two or three components.
Biology
The natural and experimental host ranges of individual viruses are usually restricted, except for a few members of the genus Crinivirus. Disease symptoms are of the discoloration type (i.e. yellowing or reddening of the leaves, small and late ripening fruit), as well as stunting, rolling, pitting and/or grooving of the woody cylinder of woody hosts. Infection is systemic, but usually limited to the phloem, which may necrotize to a varying extent. Members of a few species of the genus Closterovirus are transmissible by mechanical inoculation, though with difficulty, but no members of the genera Ampelovirus, Crinivirus and Velarivirus are mechanically transmissible. In vegetatively propagated crops, long-distance virus dissemination is primarily through infected propagating material. Members of some species can be transmitted through dodder (Cuscuta spp.). Transmission through seeds has not been proven. According to the genus, natural vectors are aphids, whiteflies, pseudococcid mealybugs and soft scale insects. Transmission is semi-persistent regardless of the type of vector. Virus geographical distribution varies from restricted to widespread, depending on the species, mostly in temperate or subtropical regions. Virions are usually found in the phloem (sieve tubes, companion cells and phloem parenchyma), occasionally in the mesophyll and epidermis. Ultrastructural modifications arise by membrane proliferation, degeneration and vesiculation of mitochondria, and formation of inclusion bodies. These are made up of aggregates of virions or membranous vesicles, or a combination of the two. Virions accumulate in conspicuous cross-banded fibrous masses or, more typically, in more or less loose bundles intermingled with single or clustered membranous vesicles. Inclusions of this type are one of the hallmarks of infection by members of the family. The vesicles contain a fibrillar network and derive either from the endoplasmic reticulum, or from peripheral vesiculation of mitochondria. Virus-encoded polyproteins 1a and 1b direct membrane remodeling and formation of multi-vesicular replication factories. Polyprotein 1a of BYV contains a variable central region between the Mtr and Hel domains (aa 1,368–1,432) which induces the formation of 1 μm mobile globules originating from endoplasmic reticulum membranes. Part of the central region is conserved in all members of the genus Closterovirus and contains a predicted amphipathic helix. This region may be involved in the biogenesis of closterovirus replication factories (Gushchin et al., 2017). The LIYV-encoded p26 localizes to plasmadesmata and is involved in systemic plant infection (Qiao et al., 2018)(Qiao et al., 2018). Members of some species in the family Closteroviridae have been engineered as gene expression and RNA interference vectors (Dawson et al., 2015, Dolja and Koonin 2013, Kurth et al., 2012).
Antigenicity
Virion proteins are moderately antigenic. Members of most virus species within a particular genus are serologically unrelated or distantly related to one another. No intergeneric serological relationship has been detected.
Derivation of names
Ampelo: from Greek ampelos, meaning grapevine, the host of members of the type species of the genus Ampelovirus.
Clostero: from Greek kloster, meaning spindle or thread.
Crini: from Latin crinis, meaning hair, from the appearance of the very long thread-like particles.
Velari: from Latin velare, meaning cryptic or veiled, because members of the type species of the genus Velarivirus do not cause any apparent disease symptoms upon infection of their natural host
Genus demarcation criteria
Members of different genera are distinguished by multiple properties (Table 2. Closteroviridae):
- Phylogenetic relationships in the RdRP, CP, HSP70h amino acid sequence
- Vector
- Number of genomic RNAs
- Number and organization of ORFs
- Virion length
Table 2. Closteroviridae. Distinguishing properties of the viruses belonging to four genera in the family Closteroviridae.
| Genus | Virion length (nm) | Number of genomic RNA species | Genome size (kb) | ORF (No.) | Replicase (kDa) | HSP70h (kDa) | CP (kDa) | CPm (kDa) | Vector |
|---|---|---|---|---|---|---|---|---|---|
| Closterovirus | 1,350–2,000 | 1 | 14.5–19.3 | 8–12 | 349–367 | 65–67 | 22–25 | 24–27 | Aphids |
| Ampelovirus | 1,400–2,200 | 1 | 13.0–18.5 | 7–12 | 245–293 | 57–59 | 28–36 | 50–56 | Mealybugs, soft scales |
| Crinivirus | 650–850 and 700–900 | 2 or 3 | 15.6–17.5 | 9–13 | 267–280 | 62–65 | 28–29 | 53–80 | Whiteflies |
| Velarivirus | 1,500–1,700 | 1 | 16.2–16.9 | 9 | 260–270 | 62–69 | 34–46 | 69–76 | None known |
Relationships within the family
Phylogenetic relationships within the family are depicted in Figure 4. Closteroviridae. Members of the family group in four distinct genera (Martelli et al., 2012).
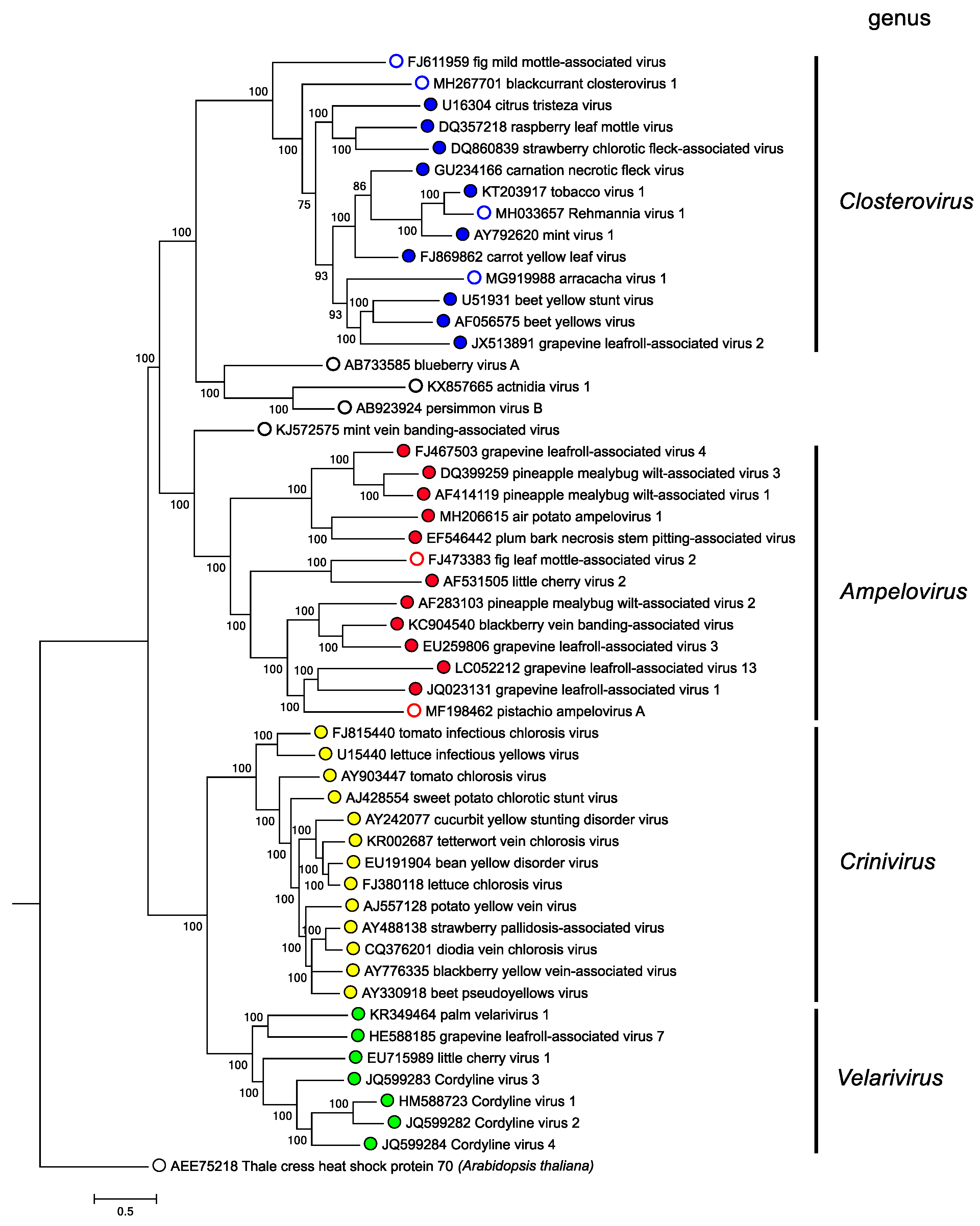 |
| Figure 4. Closteroviridae. Phylogenetic tree showing the relationships between the species and genera of the family Closteroviridae based on the amino acid sequence of the HSP70h gene. The maximum-likelihood tree was produced and bootstrapped using MUSCLE and inferred using RAxML in the T-REX web server. Branch lengths are proportional to sequence distances, and the bar represents the genetic distance. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Virions of members of some genera in the families Alphaflexiviridae and Betaflexiviridae have a similar particle morphology as those of members of the family Closteroviridae, although tending to be somewhat shorter. However, the CP amino acid sequences of members of these families are very different. Major differences also exist in genome length and organization, as well as in their strategy of expression. Replication-associated proteins (RdRP, Mtr and Hel) contain signature sequences homologous to those of other taxa of the "alpha-like" supergroup of positive-sense ssRNA viruses, the closest affinity being with members of the families Bromoviridae and Virgaviridae. The replication strategy, based on polyprotein processing, translational frameshifting and multiple sgRNA generation, closely resembles that of viruses in the families Coronaviridae and Arteriviridae. However, unlike closterovirids, the RdRP of coronaviruses and arteriviruses belong to the "picorna-like" supergroup of polymerases. Hence, the transcriptional strategy of members of the family Closteroviridae follows the mechanism of other alpha-like viruses, and is distinct from the discontinuous, leader-primed transcription of coronaviruses and arteriviruses.

