Family: Cystoviridae
Minna M. Poranen and Sari Mäntynen
The citation for this ICTV Report chapter is the summary published as Poranen et al., (2017):
ICTV Virus Taxonomy Profile: Cystoviridae, Journal of General Virology, 98: 2423–2424.
Corresponding author: Minna M. Poranen ([email protected])
Edited by: Andrew M. Kropinski and Stuart G. Siddell
Posted: October 2017, updated September 2019, March 2022
PDF: ICTV_Cystoviridae.pdf (2019 version)
Summary
The family Cystoviridae includes enveloped viruses with a tri-segmented dsRNA genome enclosed in one or two concentric, icosahedrally-symmetric protein shells. The innermost protein layer is a polymerase complex responsible for genome packaging, replication and transcription. Cystoviruses infect Gram-negative bacteria, primarily plant-pathogenic Pseudomonas syringae strains (Table 1 Cystoviridae).
Table 1 Cystoviridae. Characteristics of members of the family Cystoviridae.
| Characteristic | Description |
| Example | Pseudomonas phage phi6 (Segment S, M12921; Segment M, M17462; Segment L, M17461), species Orthocystovirus phi6 |
| Virion | Enveloped virions (about 85 nm) with typically two concentric, icosahedrally symmetric protein layers: the nucleocapsid surface shell (T=13) and the polymerase complex core (T=1). Spikes protrude from the virion surface |
| Genome | Three segments of linear, double-stranded RNA, 12.7–15.0 kbp in total. All segments encode several viral proteins |
| Replication | Single-stranded genomic precursor molecules are packaged into the viral polymerase complex. The packaged RNA molecules are replicated and transcribed within the particle |
| Translation | Viral proteins are translated from polycistronic messenger RNAs |
| Host range | Gram-negative bacteria, mostly Pseudomonas species |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Duplornaviricota, class Vidaverviricetes, order Mindivirales: seven genera and nine species |
Virion
Morphology
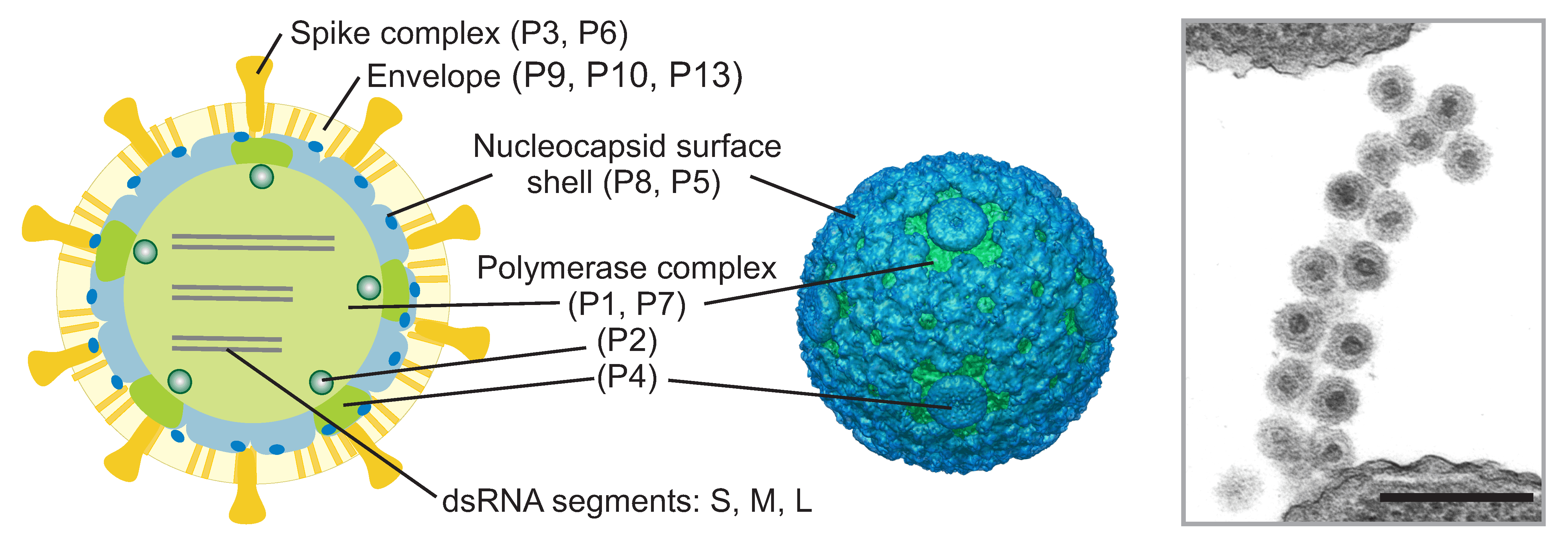
The enveloped virions are spherical, about 85 nm in diameter and covered by spikes (Figure 1. Cystoviridae). The envelope surrounds an isometric nucleocapsid, about 58 nm in diameter. However, Pseudomonas phage phi8 is lacking the nucleocapsid shell and has a single protein shell surrounding the genome. The nucleocapsid surface shell (if present) follows T=13 isocahedral symmetry. Turret-like extrusions of the underlying polymerase complex span the nucleocapsid surface shell layer at the five-fold symmetry positions (Figure 1. Cystoviridae). The dodecahedral polymerase complex is about 50 nm in diameter. In the polymerase complex major capsid protein dimers are arranged on a T=1 icosahedral lattice (Butcher et al., 1997, Huiskonen et al., 2006, Sun et al., 2017).
Physicochemical and physical properties
The molecular mass of Pseudomonas phage phi6 virion is 99 ×106 Da and the nucleocapsid is 40 ×106 Da. Virion S20,w is about 405S. The buoyant density of the virion is 1.27 g cm−3 in CsCl, 1.22 g cm−3 in Cs2SO4 and 1.24 g cm−3 in sucrose. Virions are sensitive to detergents, ether and chloroform but stable between pH 6.0 and 9.5.
Nucleic acid
Virions contain three segments of linear, double-stranded RNA: Segment L (6.4–7.1 kbp), Segment M (3.6–4.7 kbp) and Segment S (2.3–3.2 kbp). The complete genome is 12.7–15.0 kbp and has a GC content of 53.4–58.8%. Virion particles contain a single copy of each of the three genome segments. The genome constitutes approximately 10% of the virion weight. The genome is organized into five concentric layers arranged into nearly coaxial spools (Ilca et al., 2019).
Proteins
Proteins constitute about 70% of the virion weight. The viral genome (Figure 2.Cystoviridae) encodes structural (Figure 1.Cystoviridae) and non-structural proteins. The envelope contains several integral membrane proteins, including the fusogenic protein P6 and major envelope protein P9. Receptor binding spike (P3) is anchored to the envelope via P6 (Sinclair et al., 1975, Gottlieb et al., 1988a) (Bamford et al., 1987). In some cystoviruses the spike is composed of several different polypeptides. Protein P8 forms the nucleocapsid surface shell (Figure 1.Cystoviridae) (Sun et al., 2017) or is associated with the membrane (phi8, (Hoogstraten et al., 2000, Jäälinoja et al., 2007). Protein P5 is a lytic enzyme (Mindich and Lehman 1979, Dessau et al., 2012) associated on the nucleocapsid surface. Major capsid protein P1 (120 copies per virion) forms the structural skeleton of the polymerase complex (Olkkonen and Bamford 1987). Protein P2 is the viral replicase and transcriptase and is located in the interior of the P1 shell (Ilca et al., 2015, Makeyev and Bamford 2000, Butcher et al., 2001). The turret-like extrusions of the polymerase complex are made by hexamers of P4 protein (Sun et al., 2017). P4 is a nucleoside triphosphatase required for genome packaging and transcription (Pirttimaa et al., 2002, Sun et al., 2013). Minor capsid protein P7 is an assembly factor (Poranen et al., 2001). Non-structural protein P12 is a morphogenetic protein that is probably involved in envelope assembly (Johnson and Mindich 1994). Some cystoviruses also encode other non-structural proteins.
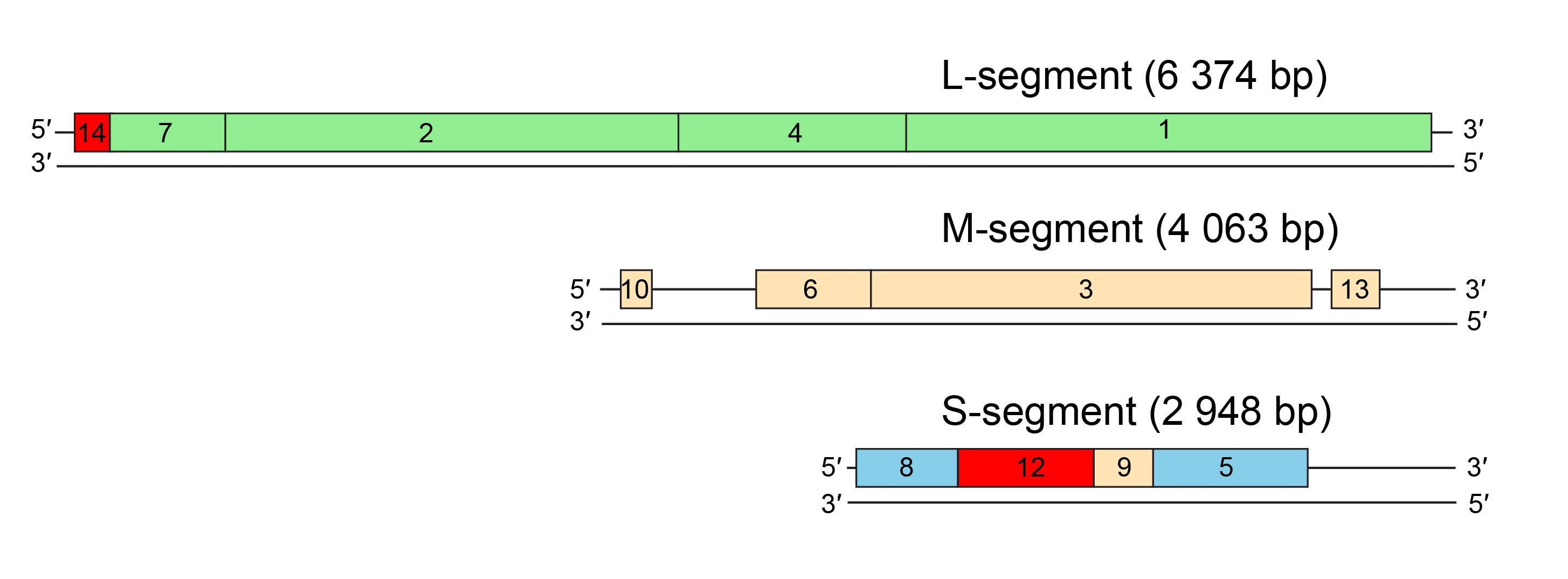 |
| Figure 2.Cystoviridae. Genome organization of Pseudomonas phage phi6. The gene and protein numbers are the same. Colourings indicate genes encoding constituents of the polymerase complex (green), nucleocapsid (blue), envelope-associated proteins (beige), and non-structural proteins (red). |
Lipids
Virions are composed of 20% lipids by weight. There is enough lipid to cover about one-half of the envelope surface area (the rest being protein). Viral lipids are derived from the host plasma membrane (Laurinavičius et al., 2007). The lipid compositions of the viral envelope and the host plasma membrane are similar.
Genome organization and replication
Distinct noncoding regions at the termini of the three genome segments contain signals for genome packaging and replication (Gottlieb et al., 1988a, Mindich et al., 1988, McGraw et al., 1986). Genes are clustered into functional groups comprising those encoding the polymerase complex, envelope-associated proteins and the nucleocapsid (Figure 2.Cystoviridae).
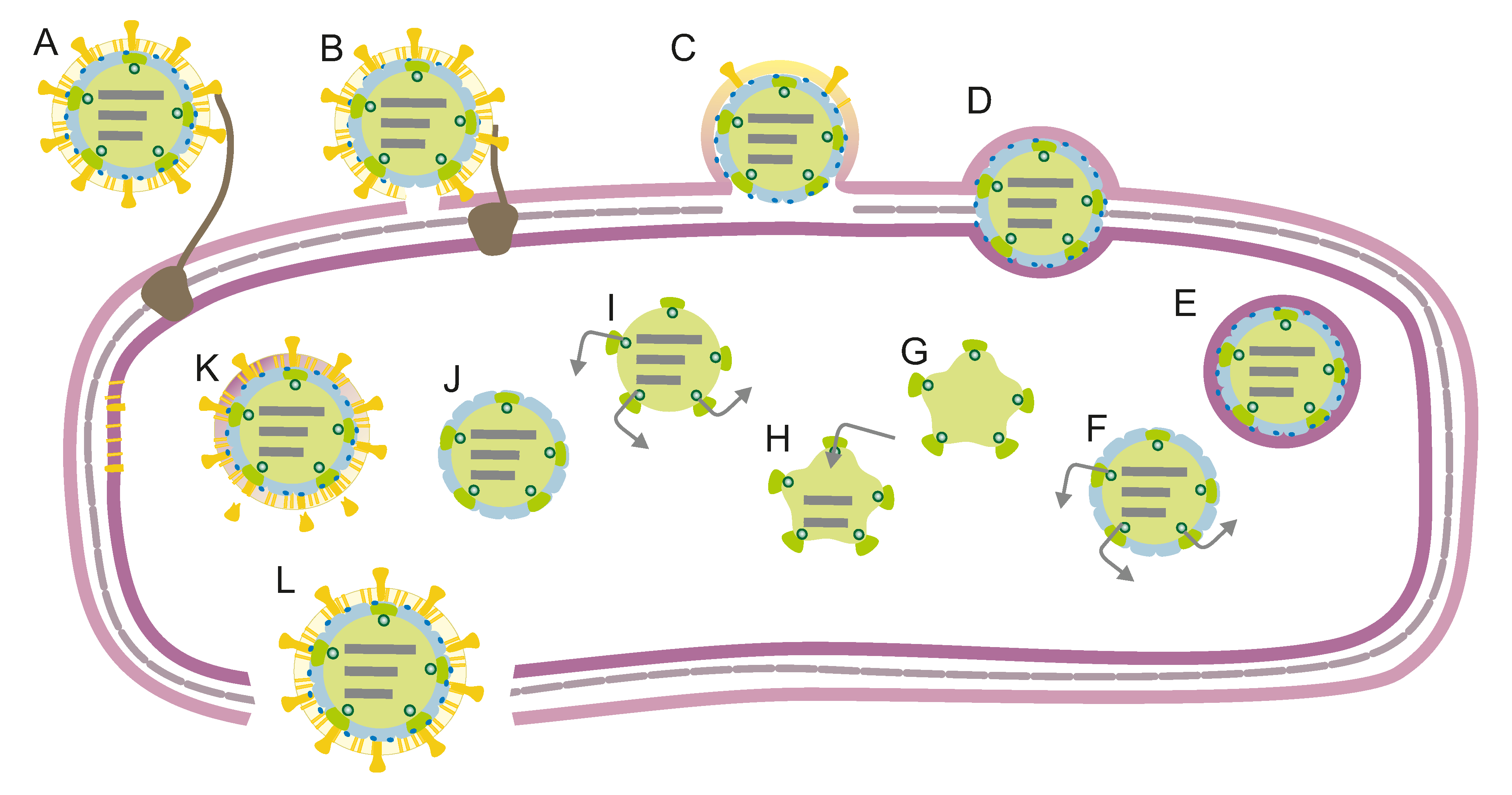
Virions adsorb either to the rough lipopolysaccharide layer or to pili of the host bacterium (Bamford et al., 1976, Roine et al., 1998, Mindich et al., 1999) (Figure 3.Cystoviridae). The viral envelope fuses with the host outer membrane (Bamford et al., 1987) and the nucleocapsid-associated lytic enzyme locally digests the peptidoglycan layer (Mindich and Lehman 1979). Delivery of the viral polymerase complex into the host cytoplasm involves an endocytic-like process at the host plasma membrane (Poranen et al., 1999). The viral genome is transcribed by the virion-associated RNA-directed RNA polymerase within the polymerase complex. Early in the infection approximately equal amounts of messenger RNA molecules are produced from the L, M and S segments (Coplin et al., 1975, Emori et al., 1983). Later in the infection cycle transcripts of the M and S segments typically predominate. Transcription is semi-conservative and produces full-length copies of the genome segments (Usala et al., 1980, Van Etten et al., 1980).
Virus mRNAs are polycistronic. Translation of L segment transcripts produces the early proteins, which assemble to form empty polymerase complexes (Figure 3.Cystoviridae) (Gottlieb et al., 1988b). These package the three positive-strand transcripts (Frilander and Bamford 1995). Negative-strand synthesis then takes place inside the polymerase complex. RNA packaging and replication induce structural changes in the polymerase complex (expansion) (Nemecek et al., 2011). Transcription by these polymerase complexes produces messages for late protein synthesis. The nucleocapsid surface shell assembles on the polymerase complex (Figure 3.Cystoviridae) (Poranen et al., 2001, Olkkonen et al., 1991). Nucleocapsid acquires protein P5 and the envelope. Spikes are assembled on the envelope surface. Mature virions are released upon virus-induced host cell lysis (Bamford et al., 1976).
Biology
Cystoviruses are lytic bacteriophages that induce host cell lysis at the end of the viral reproduction cycle. Natural hosts are Gram-negative plant or human pathogenic bacteria.
Derivation of names
Cystoviridae, Cystovirus: from the Greek kystis, “bladder”, “sack”.
Relationships within the family
Phylogenetic relationships among members of the family Cystoviridae are shown in Figure 4.Cystoviridae.
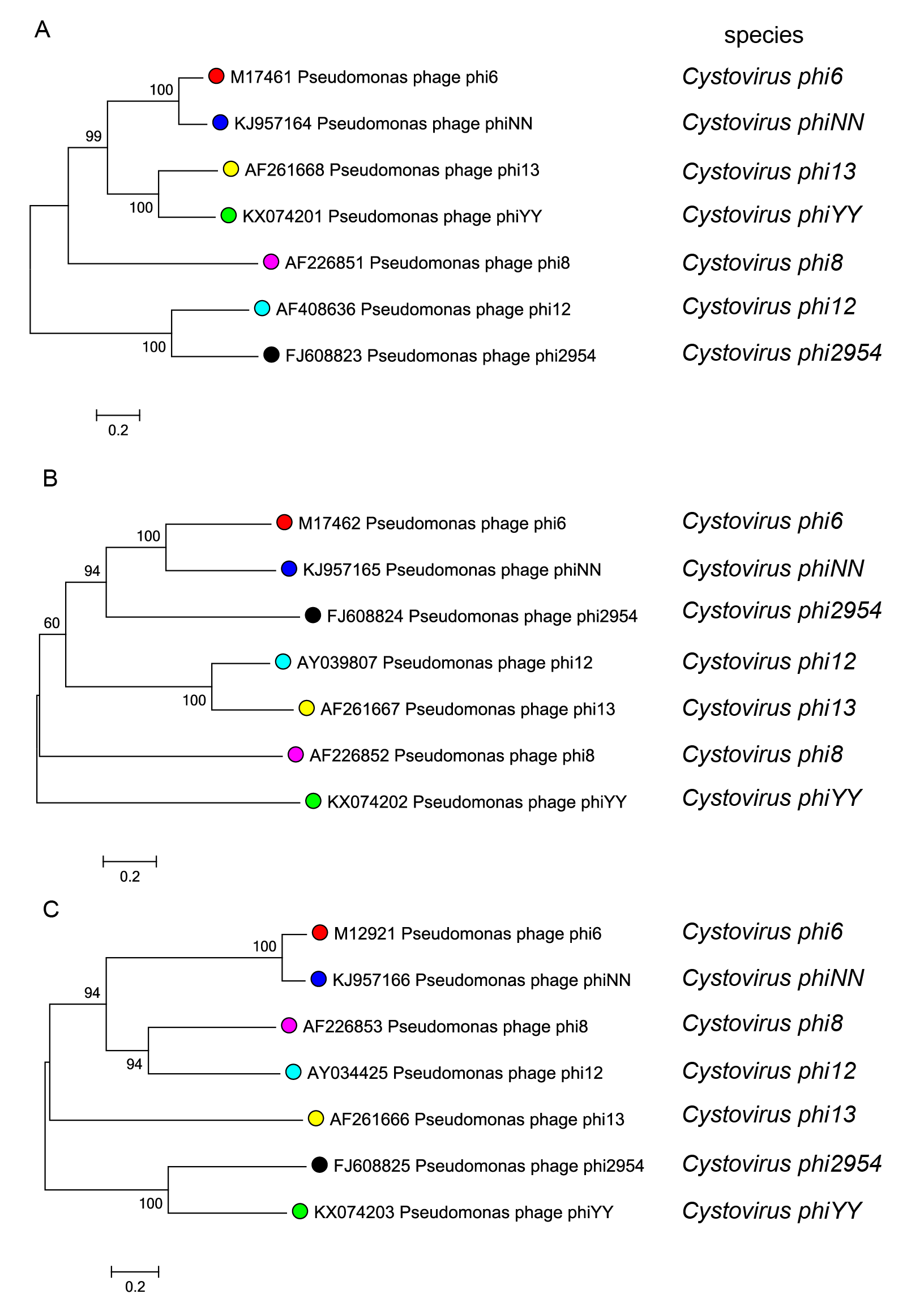 |
| Figure 4.Cystoviridae. Phylogenetic analysis of cystoviruses based on nucleotide sequence comparisons of the genome segments L (A), M (B) and S (C). Phylogenetic trees were constructed using MEGA 7.0 software (Kumar et al., 2016) with maximum likelihood method. Bootstrap values greater than 50% (1000 replicates) are shown at branch points. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
In terms of genome replication strategy, cystoviruses resemble eukaryotic double-stranded RNA viruses (Mertens 2004, Poranen and Bamford 2012). The structure, organization and functions of the polymerase complex are the major similarities among members of families Cystoviridae, Chrysoviridae, Reoviridae, Totiviridae, Partitiviridae, Megabirnaviridae, Quadriviridae and Picobirnaviridae The T=13 architecture of the surrounding capsid layer is also shared by cystoviruses and reoviruses.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Microvirgula phage phiNY | S: MW471135; M: MW471134; L: MW471133 | phiNY |
| Pseudomonas phage phi7* | S: AF125682; M: AF125681; L: AF125680 | phi7 |
| Pseudomonas phage phi9* | S: AF125679; M: AF125678; L: AF125677 | phi9 |
| Pseudomonas phage phi10* | S: AF125676; M: AF125675; L: AF125674 | phi10 |
| Pseudomonas phage phi11* | phi11 | |
| Pseudomonas phage phi14* | phi14 |
Virus names and virus abbreviations are not official ICTV designations.
*Either none, or only partial sequences are available for these isolates (Mindich et al., 1999).