Family: Finnlakeviridae
Sari Mäntynen#, Elina Laanto#, Lotta-Riina Sundberg, Minna M. Poranen and Hanna M. Oksanen
# Shared first authorship
Corresponding author: Hanna M. Oksanen ([email protected])
Edited by: Evelien Adriaenssens and Stuart G. Siddell
Posted: July 2020, updated May 2022, May 2023
PDF: ICTV_Finnlakeviridae.pdf (2020 version)
Summary
Finnlakeviridae is a family of icosahedral, internal membrane-containing bacterial viruses with circular, single-stranded DNA genomes (Table 1. Finnlakeviridae). The family includes a single genus, Finnlakevirus, with one species, Finnlakevirus FLiP. Flavobacterium phage FLiP was isolated with its gram-negative host bacterium from a boreal freshwater habitat in Central Finland in 2010. It is the first described single-stranded DNA virus with an internal membrane. Flavobacterium phage FLiP shares minimal sequence similarity with other known viruses. The virion organization (pseudo T = 21 dextro) and major capsid protein fold (double-ß-barrel) of Flavobacterium phage FLiP resemble those of Pseudoalteromonas phage PM2 (family Corticoviridae) that has a double-stranded DNA genome. A similar major capsid protein fold is also found in other double-stranded DNA viruses in the kingdom Bamfordvirae. Since only one genus is currently recognized within the family Finnlakeviridae, the genus description corresponds to the family description.
Table 1. Finnlakeviridae. Characteristics of members of the family Finnlakeviridae.
| Characteristic | Description |
| Typical member | Flavobacterium phage FLiP (MF361639), species Finnlakevirus FLiP, genus Finnlakevirus |
| Virion | Icosahedral, internal membrane-containing virions, approximately 59 nm in diameter (from vertex to vertex). Spikes protrude from the virion surface |
| Genome | 9.2 kb of circular, single-stranded DNA |
| Replication | Possibly rolling circle replication (not experimentally shown) |
| Translation | Prokaryotic translation |
| Host range | Gram-negative bacteria from the genus Flavobacterium |
| Taxonomy | Realm Varidnaviria, kingdom Abadenavirae, phylum Produgelaviricota, class Ainoaviricetes, order Lautamovirales; one genus with one species |
Virion
Morphology
The virion of Flavobacterium phage FLiP consists of an icosahedral protein shell and an internal membrane, which are uniquely combined with a circular single-stranded DNA genome (Figure 1. Finnlakeviridae). The diameter of the virion is about 59 nm (vertex-to-vertex). The inner surface of the icosahedral protein capsid is covered by a lipid bilayer membrane (5 nm thick), enclosing the single-stranded DNA genome. The major capsid proteins (MCPs) forming the outer protein shell follow a pseudo T = 21 dextro icosahedral capsid organization, previously described only for marine double-stranded DNA Pseudoalteromonas phage PM2 (Huiskonen et al., 2004). The Flavobacterium phage FLiP MCPs consist of two β-barrels with jellyroll topology and arrange into trimers to form pseudohexameric molecules. The five-fold vertices are occupied by pentameric spike complexes (Laanto et al., 2017).
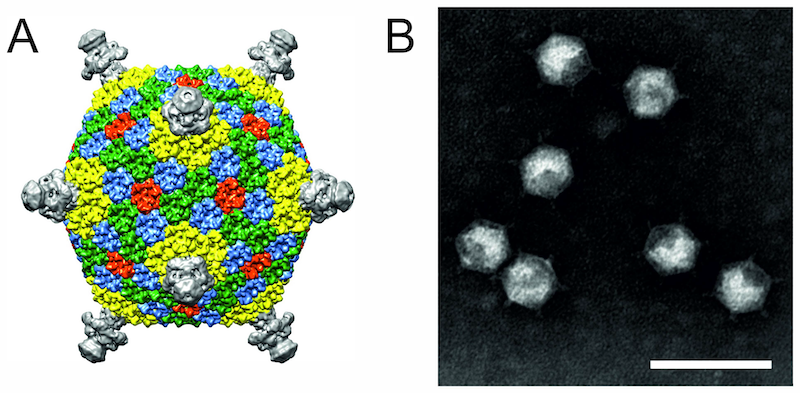 |
| Figure 1. Finnlakeviridae. (A) Cryo-electron microscopic reconstruction of the Flavobacterium phage FLiP virion (Laanto et al., 2017). (B) Flavobacterium phage FLiP virions negatively-stained with 2% phosphotungstic acid (pH 8.5) and visualized under transmission electron microscopy. Scale bar represents 100 nm. |
Physicochemical and physical properties
The virions of FLiP are sensitive to chloroform. Their buoyant density is 1.21 g/ mL in CsCl and 1.18 g/mL in sucrose (Laanto et al., 2017). Viruses are sensitive to temperature and infectivity drops one order of magnitude in a week at 22 °C.
Nucleic acid
Flavobacterium phage FLiP virions contain a single copy of a circular single-stranded DNA of 9,174 nucleotides and the GC content of the genome is 34% (Laanto et al., 2017).
Proteins
The viral genome contains 16 predicted coding sequences (CDSs), five of which have been shown to encode structural proteins (P7-P9, P11, P14; Figure 2. Finnlakeviridae). The MCP P8 (encoded by CDS8) is the most abundant protein in the virion. CDS14 shares a significant sequence similarity with several lytic transglycosylases, suggesting that P14 is a lytic protein present in the virion (Laanto et al., 2017) and likely assisting in the penetration of peptidoglycan layer during entry.
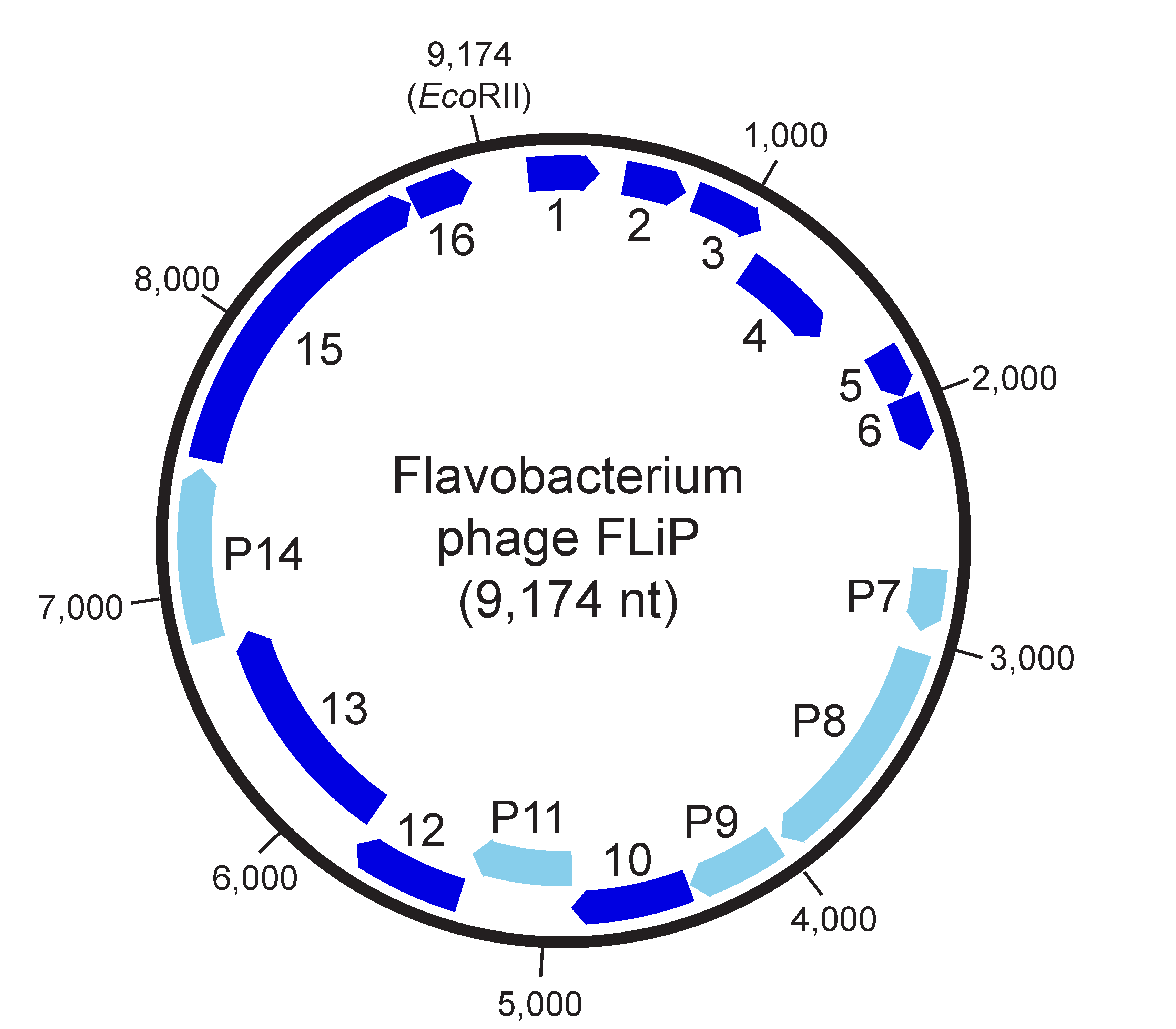 |
| Figure 2. Finnlakeviridae. Genome organization of Flavobacterium phage FLiP. Nucleotide numbering starts from a unique EcoRII restriction site. Sixteen predicted coding sequences are marked with arrows indicating the direction of transcription. CDS encoding structural proteins are indicated by light blue colour and labelled with the protein they encode. |
Lipids
Lipids form an internal membrane that also contains virus-encoded proteins. The major lipid class of the virion membrane is ceramide (60 % of the total lipid composition). The viral lipid composition differs from that of the host cytoplasmic membrane indicating selective uptake of the lipids during virion assembly (Laanto et al., 2017).
Genome organization and replication
All of the 16 predicted CDSs are oriented in the same direction (Figure 2. .Finnlakeviridae). They show a limited sequence identity to other sequences in the public databases. Five of the CDSs encode structural proteins (CDS7-9, 11, 14). CDS14 shares homology with lytic enzymes. Flavobacterium phage FLiP does not encode any identifiable DNA or RNA polymerase. Sequence resemblance of CDS15 to rolling circle replication initiator proteins, and especially to replication proteins of filamentous M13-like inoviruses, suggests a replication mechanism typical of single-stranded DNA phages (Laanto et al., 2017, Yutin et al., 2018). The absence of a putative packaging ATPase implies that a viral-encoded ATPase is not required for the encapsidation of the Flavobacterium phage FLiP genome (Yutin et al., 2018).
Biology
Flavobacterium phage FLiP is a virulent virus that induces host cell lysis at the end of the viral reproduction cycle. The isolation host of Flavobacterium phage FLiP is a member of genus Flavobacterium (Laanto et al., 2017). Flavobacterium species are commonly found in boreal freshwater habitats (Bernardet and Bowman 2006).
Derivation of names
Finnlakeviridae: from English Finn (Finnish) and from English lake in reference to the isolation locality
Relationships with other taxa
Flavobacterium phage FLiP shows only limited sequence identity to other known viruses. However, Flavobacterium phage FLiP shares striking structural similarities with Pseudoalteromonas phage PM2 (family Corticoviridae), both in terms of the unique capsid organization (pseudo T = 21 dextro) and the major capsid protein fold (double-ß-barrel) (Laanto et al., 2017, Abrescia et al., 2008). Similar major capsid protein folds can be found in members of the kingdom Bamfordvirae (Koonin et al., 2020), such as Enterobacteria phage PRD1 (family Tectiviridae) (Benson et al., 1999), archaeal virus Sulfolobus turreted icosahedral virus 1 (family Turriviridae) (Maaty et al., 2006), eukaryotic viruses Paramecium bulsaria Chlorella virus 1 (family Phycodnaviridae) (Nandhagopal et al., 2002), Chilo iridescent virus (family Iridoviridae) (Yan et al., 2009) and human adenovirus (family Adenoviridae) (Rux et al., 2003), as well as in Sputnik virophage (a satellite virus of Mimivirus, family Lavidaviridae) (Zhang et al., 2012). Based on phylogenetic analysis of the MCP sequences, Flavobacterium phage FLiP forms its own group among the tailless prokaryotic DNA viruses and proviruses with double-ß-barrel MCPs (Yutin et al., 2018).

