Family: Hepadnaviridae
Lars Magnius, William S. Mason, John Taylor, Michael Kann, Dieter Glebe, Paul Dény, Camille Sureau and Helene Norder
The citation for this ICTV Report chapter is the summary published as Magnius et al., (2020):
ICTV Virus Taxonomy Profile: Hepadnaviridae, Journal of General Virology, 101:571–572.
Corresponding author: Lars Magnius ([email protected])
Edited by: Balázs Harrach and Peter Simmonds
Posted: March 2020
PDF: ICTV_Hepadnaviridae.pdf
Summary
Hepadnaviridae is a family of small enveloped viruses with partially double-stranded DNA of approximately 3.2 kb (Table 1. Hepadnaviridae). Common features of all members of the family are the expression of three major sets of proteins, (precore/core,polymerase,preS/S) and replication by reverse transcription within immature nucleocapsids in the cytoplasm of infected hepatocytes. Hepadnaviruses are hepatotropic and infections may be transient or persistent. There are five genera, Parahepadnavirus, Metahepadnavirus, Herpetohepadnavirus, Avihepadnavirus, and Orthohepadnavirus. Members of both Metahepadnavirus and Parahepadnavirus infect teleost fish, and the majority of viruses in these genera have been identified by metagenomic approaches. Only white sucker hepatitis B virus (Parahepadnavirus) and bluegill hepatitis B virus (Metahepadnavirus) have been found as virions in infected hosts. Members of the genus Herpetohepadnavirus infect reptiles and frogs. The genus Avihepadnavirus comprises three species whose members infect birds and the genus Orthohepadnavirus comprises 12 species whose members infect mammals. The family is placed within the realm Riboviria, order Blubervirales, because of homology between the hepadnavirus reverse transcriptase and the RNA-directed RNA polymerase of RNA viruses (Koonin et al., 2020).
Table 1. Hepadnaviridae. Characteristics of members of the family Hepadnaviridae
| Characteristic | Description |
| Typical member | hepatitis B virus, genotype D (V01460), species Orthohepadnavirus hominoidei |
| Virion | Envelope of 42–50 nm diameter surrounding a nucleocapsid usually composed of 240 protein subunits |
| Genome | 3.0–3.4 kb partially double-stranded DNA |
| Replication | Pre-genomic RNA transcripts from covalently closed in the nucleus are encapsidated and reverse-transcribed in the cytoplasm |
| Translation | Five or six mRNAs with different 5′-ends and a common 3′-end linked to a polyadenylation site |
| Host range | Teleost fish (Parahepadnavirus and Metahepadnavirus), reptiles and frogs (Herpetohepadnavirus), birds (Avihepadnavirus), mammals (Orthohepadnavirus), |
| Taxonomy | Realm Riboviria, kingdom Pararnavirae, phylum Artverviricota, class Revtraviricetes, order Blubervirales , five genera with 27 species |
Virion
Morphology
Hepatitis B virus (HBV) is spherical, occasionally pleomorphic, 42–50 nm in diameter, with no evident surface projections after negative-staining. The outer, detergent-sensitive, envelope contains two or three surface proteins, which suffice to induce protective immunity (Bruss 2007), and surrounds an icosahedral nucleocapsid (Bruss 2007). The nucleocapsid, also termed core, is composed of one major protein, the core protein. It encloses the viral partially double-stranded DNA genome, the viral DNA polymerase, and cellular proteins, including a protein kinase and chaperones, all acting in the initiation of viral DNA synthesis.
For HBV, the majority of nucleocapsid cores are about 36 nm in diameter and contain 240 core protein subunits (triangulation number T=4), while a minority are approximately 32 nm in diameter and consist of only 180 subunits (T=3) (Beterams et al., 2000).
Hepadnaviruses induce the overproduction of surface proteins that are secreted into the blood as pleomorphic particles. For HBV, these are 17–22 nm spherical particles and filaments (Figure 1. Hepadnaviridae). Virions and empty particles contain two or three surface proteins, each in more than one form due to alternative glycosylation.
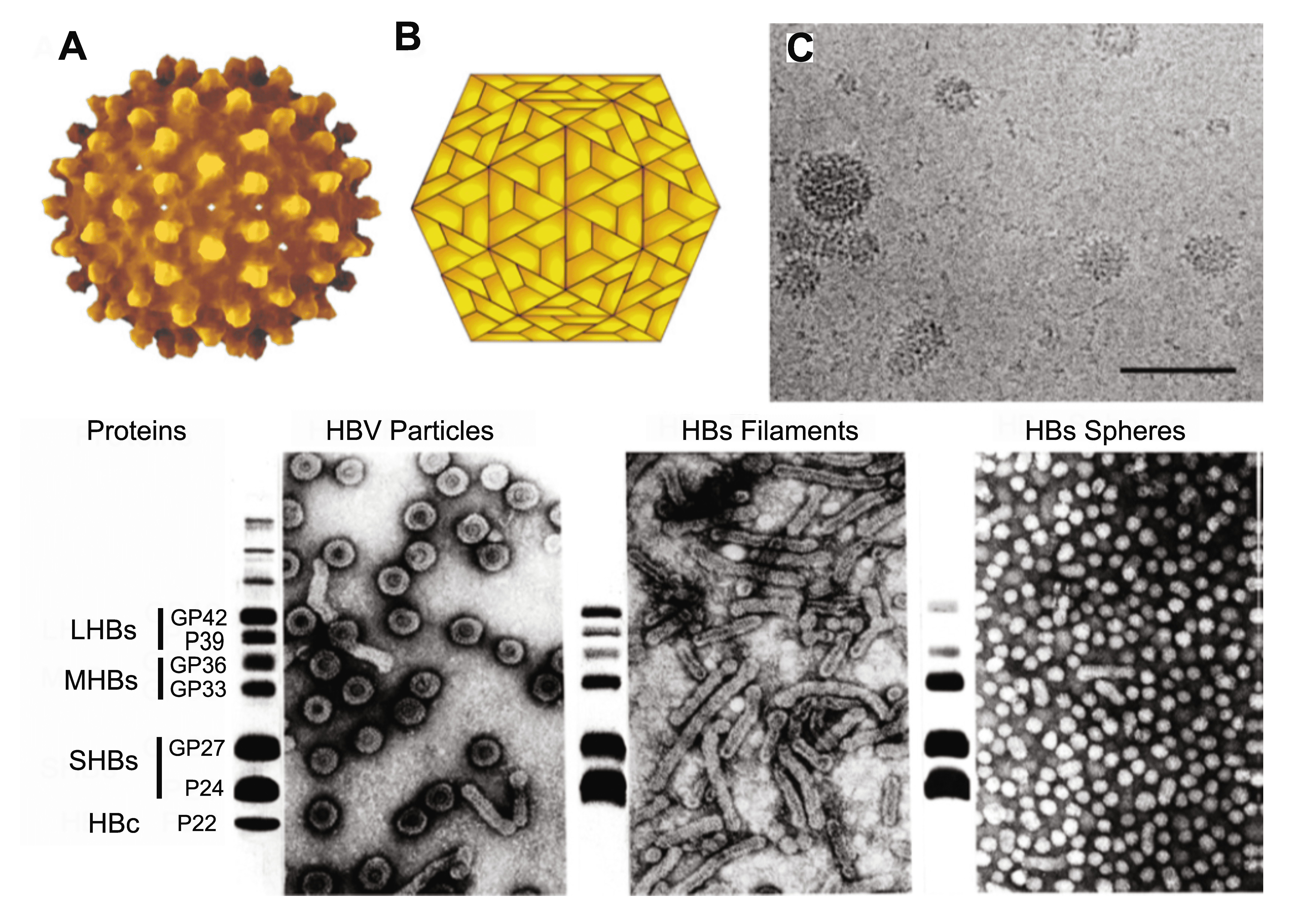 |
| Figure 1. Hepadnaviridae. (A) Atomic resolution rendering of a particle of hepatitis B virus (HBV) capsid (Rasmol (Sayle and Milner-White 1995) image from PDB 1qgt (Wynne et al., 1999) courtesy of Dr. J.-Y. Sgro, UW-Madison, USA, http://www.virology.wisc.edu/virusworld). (B) Diagram representing the T=4 structure of an HBV core particle. (C) High resolution cryo-electron micrograph of normal (42–52 nm) isometric virus and of smaller (ca. 22 nm) spheres and rods composed of viral envelope proteins. The bar represents 65 nm. (Courtesy of B. Boettcher, J. Monjardino and R.A. Crowther.) (Bottom). Negative-contrast electron micrographs of HBV virions (left) and virus-associated particles (centre and right), together with an SDS-PAGE protein profile of each particle form to the left of the relevant micrograph. LHBs, MHBs and SHBs refer to large, middle and small HB surface proteins, respectively. HBc, hepatitis B core proteins. GP, glycoprotein; P, protein. The identities of the slower migrating bands are unknown. (Courtesy of W. Gerlich.) A recent report suggests that the majority of HBV particles, presumably those with a dark center as the result of accumulation of stain, are devoid of nucleic acids and are non-infectious (Ning et al., 2011). |
Physicochemical and physical properties
The virion S20,w is about 280S. The buoyant density of viral DNA containing virions in CsCl is 1.25 g cm−3. Estimates of the buoyant density of particles lacking cores are 1.18–1.20 g cm−3. Virus-derived cores (lacking envelopes but containing nucleic acid) have densities of approximately 1.36 g cm-3 (Hruska and Robinson 1977).
Nucleic acid
The hepadnavirus genome consists of a partially dsDNA held in a circular conformation by base pairing in a cohesive overlap between the 5′-ends of the two DNA strands (Seeger and Mason 2015) (Figure 2. Hepadnaviridae). The length of the cohesive overlap is about 240 bp for orthohepadnaviruses and 50 bp for avihepadnaviruses. The genome length ranges from 3.0 to 3.4kb between members of species belonging to different genera; the viral DNA has an S20,w of about 14S and a G+C content of about 48%. One strand (negative-sense, i.e. complementary to the viral mRNAs) is full-length, whereas the other varies in length. The negative-sense strand DNA has an 8–9 nt terminal redundancy. The 5′-end of the negative-sense strand DNA is covalently attached to a tyrosine residue in the terminal protein (TP) domain of the viral DNA polymerase, and the 5′-end of the positive-sense DNA has a covalently attached 19 nt, 5′-capped oligoribonucleotide primer. The 3′-end of the positive-sense strand terminates at a variable position in different molecules, creating a single-stranded gap that may account for as much as 60% of the HBV genome.
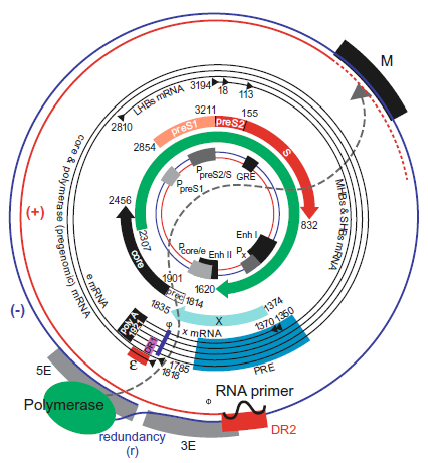 |
| Figure 2. Hepadnaviridae. Genome organization and regulatory elements of orthohepadnaviruses are shown for a typical HBV isolate of genotype A. The outer circle represents the structure of relaxed circular, viral DNA found within virions, while the inner circle illustrates the structure and regulatory elements on cccDNA, the covalently closed circular DNA from which viral mRNAs are transcribed in the nucleus of the infected cell (red = positive-sense strand; blue = negative-sense strand). Numbering starts at the unique EcoRI restriction site located approximately at the junction of the preS1 and preS2 domains in the ORF for the viral envelope proteins. The regulatory elements on the DNA are depicted at their approximate positions. The promoters (P) are shown as grey boxes, and the enhancers (Enh), a glucocorticoid responsive element (GRE), and a CCAAT element (CCAAT) in the preS2/S promoter are depicted as black boxes. The basal core promoter is regulated by the negative regulatory element (NRE, not shown), which overlaps with Enh II. Liver-specific promoters are drawn in light grey; non-tissue-specific promoters are depicted as medium grey boxes. The ORFs are drawn as arrows with their corresponding start and termination sites. The viral mRNAs are depicted as black circles in the middle region. The black triangles represent their 5′-ends; the 3′-end is common and linked to an approximately 300 nt poly(A) tract. The regulatory elements on the RNAs are depicted as a red box (encapsidation signal ε), a black box (polyadenylation signal), in pink (DR1), in blue (phi) and in light blue (posttranscriptional regulatory element [PRE]). The genomic DNA is depicted as it is found in the virion. The negative-sense DNA strand is drawn as a blue line with its terminal redundancy (r). The polymerase (green oval) is linked to the 5′-end of the negative-sense strand. The positive-sense strand DNA is shown as a red line. The dotted red line represents the variation of the 3′-end of the positive-sense strand DNA. The 5′-end of the positive-sense strand is bound to its capped RNA primer, depicted as a black, wavy line. The dotted grey line between the polymerase and the 3′-end of the positive-sense strand DNA reflects the fact that the polymerase is bound to the 5′-end of the negative-sense strand DNA but interacts with the variable 3′-end of the positive-sense strand DNA for its elongation. The regulatory elements on the negative-sense strand DNA are the DR2 (red box) and the M, 5E and 3E elements, which are required for circularization of the genome. Note that their position and size are approximate, since these elements are not yet completely characterized. (From (Kann 2002); with permission.) |
The genome has three open reading frames (ORFs); precore/core (preC/C), polymerase (P), env or surface (preS/S), and, regarding orthohepadnaviruses, an additional X ORF (Figure 2. Hepadnaviridae). Genomic and sub-genomic RNAs are transcribed by RNA polymerase II into mRNAs with varying 5′-ends but a common 3′-end (Seeger and Mason 2015).
Proteins
Virions and empty subviral particles may contain two or three surface proteins, with a common C-terminus but distinct N-termini due to different sites of translation initiation (Bruss 2007). Typically, virions of orthohepadnaviruses contain a small transmembrane surface protein (SHBs), an intermediate sized (MHBs) and a large (LHBs) protein that is myristoylated at the N-terminus. More than one form of each of the above proteins occurs due to alternative patterns of glycosylation. For HBV and woodchuck hepatitis virus (WHV), virions and filaments are enriched in the LHBs proteins. The empty spheres consist predominantly of S proteins. Of note, duck hepatitis B virus (DHBV, Avihepadnavirus) particles contain only LHBs and SHBs proteins, which are distributed evenly between particle types.
The core protein has a large N-terminal domain of approximately 140 aa, which is the structural component of the nucleocapsid. The 43–45 aa-long C terminus harbors a small RNA-binding domain with four arginine clusters, which comprises RNA- and DNA-binding domains, a nuclear localization signal, an importin β-binding domain, and serine phosphorylation sites. Core protein can self-assemble via dimers and subsequent hexamer formation to produce nucleocapsids in the absence of other viral or cellular components. However, interaction with nucleic acids enhances the nucleocapsid assembly.
In avi- and orthohepadnaviruses the core ORF also encodes a secreted soluble protein (HBe-antigen) that is translated from an additional start codon, 29 codons upstream and in frame with the core start codon. This additional precore sequence functions as a signal peptide, translocating the primary translation product into the ER. Most of precore is cleaved off but the remaining eight amino acids prevent assembly into a nucleocapsid-like structure. Following the secretory pathway, the modified protein is then further C terminally cleaved by furin proteases in the Golgi compartment prior to secretion. The HBe-antigen is not essential for virus replication in vivo and in vitro. Yet it is highly conserved and found in sera and seems to modulate the immune response. HBeAg negative variants arising from pre-core stop mutations or promoter mutations are often associated with a severe outcome of acute hepatitis B in their contact cases. The genomes of herpeto- and parahepadnaviruses have a region that may encode for a protein upstream and in frame with the core region, although a protein corresponding to the HBeAg has not been shown.
The polymerase protein (P protein) consists of an N-terminal domain (terminal protein, TP), a spacer region of variable size, a reverse transcriptase and an RNase H domain. The TP domain is covalently attached to the first nucleotide of 5′-end of the negative-sense strand of viral DNA via a tyrosine residue, which serves as the primer for initiation of reverse transcription.
Orthohepadnaviruses contain a fourth ORF (“X” gene) situated downstream from the S gene and partly overlapping the cohesive 5′-terminal region. This ORF codes for a non-structural protein that can function as a pleiotropic transcriptional activator and also functions in transcription of viral covalently closed circular (ccc) DNA (cccDNA). This protein is required for efficient in vivo replication of WHV. It has, however, been shown to be dispensable for replication in dedifferentiated cells, but essential for HBV replication in the differentiated hepatoma cell line HepaRG. At high expression levels in cell culture systems, the X proteins induce apoptosis. Avihepadnaviruses have an ORF in a similar location, but the corresponding protein seems to be not essential for infection. This protein has not been identified in members of the other hepadnavirus genera.
Host proteins contained within nucleocapsids include, at least, the heat shock proteins Hsp70 and Hsp90, which, in the case of DHBV, appear to be part of a multicomponent chaperone complex involved in replication and nucleocapsid assembly. A protein kinase has been detected in both ortho- and avihepadnaviral nucleocapsids. The viruses of other genera have been less investigated in this regard.
Lipids
Lipid constitutes 30–40% of the viral envelope or of the empty particles. The viral envelope is derived from a host membrane compartment intermediate between the ER and Golgi, and includes phospholipids, cholesterol, cholesterol esters, and triglycerides.
Carbohydrates
N-linked glycans of complex types are demonstrated in particles and virions of orthohepadnaviruses. Many virus isolates also contain O-linked glycans in the preS2 domain of the MHBs surface protein.
Genome organization and replication
Hepadnavirus genomes are relatively small and include overlapping coding regions and regulatory elements. The HBV genome has two enhancer regions (EnhI and EnhII), one negative regulatory element (NRE; not shown on Figure 2. Hepadnaviridae), four promoters (Pcore/e, PpreS1, PpreS2/S and PX), two 11-base direct repeat sequences (DR1 and DR2), a polyadenylation signal (TATAAA), and a putative glucocorticoid-responsive element (GRE). The 5′-end of the negative-sense strand is located within DR1 and the 5′-end of the positive strand is at the 3′-boundary of DR2.
Genomic and sub-genomic RNAs (sgRNAs) are transcribed by host RNA polymerase II into a number of RNA size classes, some of which also show microheterogeneity at the 5′-end but all terminate at a common 3′-polyadenylation site. The RNAs of HBV and WHV contain a post-transcriptional regulatory element (PRE), which allows for cytoplasmic transport without splicing. The PRE is probably present in all hepadnavirus genomes, since they all replicate via a more-than-genome length RNA transcript in the cytoplasm. The largest of these RNAs is translated to form precore protein. A slightly shorter RNA (the pregenomic RNA) encodes the core protein and (by internal initiation) the polymerase protein, and also as mentioned serves as the template for reverse transcription. The polymerase protein, along with host chaperone proteins, associates with a specific encapsidation signal (ε), which is close to the 5′-end of the pregenomic RNA. For efficient negative-sense strand DNA synthesis epsilon has to interact with the 19 nt long phi signal, which is located 32 nt upstream of DR1 at the 3′-end of the RNA (Tang and McLachlan 2002). This pre-assembly complex apparently facilitates the assembly of core protein dimers into complete nucleocapsids.
The HBe-antigen and core protein are translated from genomic, terminally redundant, polyadenylated 3.5 kb transcripts with slightly different 5′-ends. Only the longer precore mRNAs contains the preC initiation codon. As mentioned, the shorter mRNA is bicistronic, not only coding for core but also for the P protein. The corresponding P ORF located down-stream of the C ORF overlaps with the 3′-portion of the C ORF. The P ORF covers some 80% of the genome and its translation is via internal initiation.
The surface gene consists of three in-phase ORFs, termed in 5′- to 3′-direction, preS1, preS2 and S. The S ORF can be expressed separately to give the small or SHBs protein; co-translation of preS2/S yields the middle or MHBs protein (orthohepadnaviruses), and that of the entire preS1/preS2/S gene the large or LHBs protein. Thus, the S encoded protein region is common to all three forms of surface proteins. This is achieved by the generation of mRNAs with staggered 5′-ends in which the initiator codons of the preS1, the preS2 or the S region are the first to be encountered by translating ribosomes. L protein is translated from a 2.4 kb mRNA, and M and S from a set of 2.1 kb transcripts. All viral transcripts are 3′-terminally colinear, ending after a unique polyadenylation signal located in the C gene. The different gene products (LHBs, MHBs, and SHBs proteins) oligomerize and bud into the lumen of a post ER/pre-Golgi compartment to give rise to both empty subviral particles and virions.
For orthohepadnaviruses, there is evidence that the infectious HBV DNA-containing virions first bind unspecifically to the target cell via heparan-sulfate-proteoglycans and the virion S domain-interaction, followed by specific interaction of the preS1 domain of the LHBs protein to a hepatocyte receptor, sodium taurocholate co-transporting polypeptide (NTCP) that is expressed on the basolateral membrane of hepatocytes (Yan et al., 2012). Transfection with cDNA encoding NTCP confers HBV susceptibility to human cell lines that are otherwise refractory to virus entry. Following virus uptake in vesicles, the nucleocapsid reaches the cytoplasm without undergoing significant acidification. Retrograde transport to the nuclear periphery is mediated by cytoplasmic dynein and microtubules. The capsid passes the nuclear pore intact and releases the genome to the nucleoplasm. Repair of the genome requires filling of the single stranded gap by host DNA polymerases, removal of the TP domain of the virus polymerase and oligoribonucleotide from the negative- and positive-sense strands, removal of the terminal redundancies and DNA ligation. The resulting covalently closed circular DNA (cccDNA) is a histone-associated minichromosome, to which core molecules and the X protein bind. cccDNA provides a stable template for transcription.
Replication can be considered in two stages: an incoming or afferent arm in which the input viral genome enters the nucleus and is converted to cccDNA, and an outgoing or efferent arm in which RNA transcripts from the cccDNA are encapsidated and reverse-transcribed within immature core particles in the cytoplasm.
The reverse transcription of the pregenomic RNA initiates with the copying of 4 nt from a bulge in the ɛ region and uses the TP domain of the polymerase as primer for first strand synthesis. This product is then annealed to a complementary sequence at the 3′-copy of DR1, and it is from this site that synthesis of the full-length negative-sense strand progresses. Synthesis of both strands requires strand transfer reactions. Positive-sense strand synthesis involves a transfer of an RNA primer derived from the 5′-end of the template RNA and extending through the proximal copy of DR1 to a remote site, DR2, which is near the 5′-end of the negative-sense strand and identical in sequence to DR1. It is here that positive-sense strand synthesis normally begins. Positive-sense-strand elongation requires a second translocation, from the 5′- to the 3′-end of the negative-sense strand template, to form an open circular genome with a less than full-length positive-sense strand DNA, maintained by overlapping cohesive ends.
Nucleocapsids containing partly reverse transcribed DNA that have associated with cytoplasmically located preS domains of the LHBs envelope protein may then bud into the lumen of multivesicular bodies as maturing virions. Nucleocapsids of avihepadnavirus have been shown to also be transported to the nucleus, thereby increasing the pool of cccDNA, a process that has been postulated also for orthohepadnaviruses (Figure 3. Hepadnaviridae).
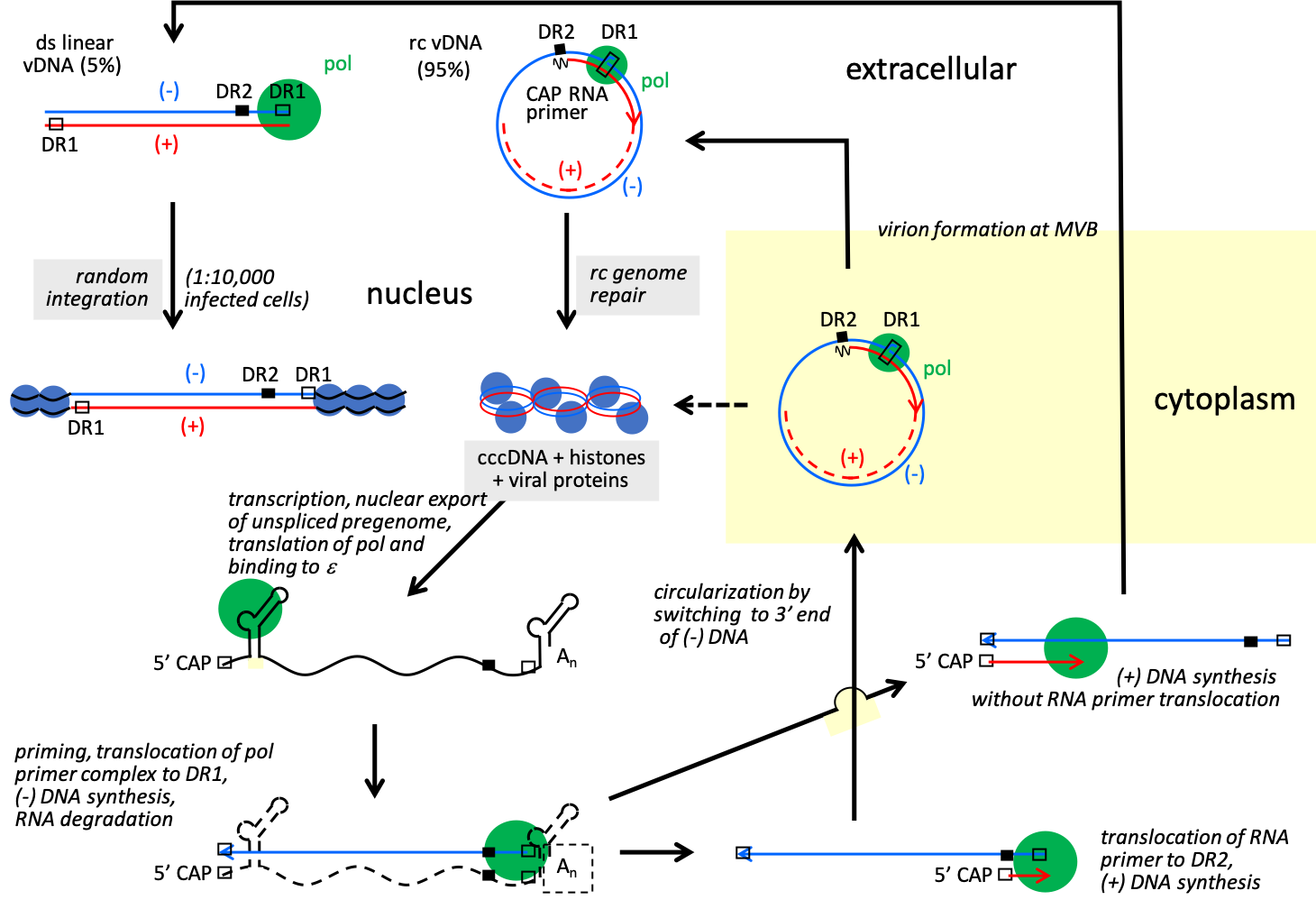 |
| Figure 3. Hepadnaviridae. Hepadnavirus replication cycle. The relaxed circular genome (rc vDNA) is repaired and as cccDNA is the template for viral mRNAs and pregenomic RNA that is encapsidated together with viral polymerase in immature core particles in the cytoplasm and reverse transcribed into rc vDNA. Reverse transcription initiates with the copying of 4 nt from a bulge in ɛ. This product is then annealed to a complementary sequence at the 3′-copy of direct repeats 1 (DR1), and it is from this site that synthesis of the full-length negative-sense strand progresses. Positive-sense strand synthesis involves a transfer of the RNA primer from the 3′-end of the negative-sense strand to a remote site, DR2, which is near the 5′-end of the negative-sense strand and identical in sequence to DR1. The core particle with rc vDNA is either transported to the nucleus thereby increasing the pool of cccDNA or enveloped and secreted. Hepadnaviruses have mostly rc vDNA, though a small fraction of viruses has linear genomes, created when the positive-sense strand RNA primer fails to translocate to DR2 prior to initiation of positive-sense strand synthesis. The linear genomes have no known role in virus replication. Upon infection, the ends of some of these linear DNAs recombine via non-homologous recombination to form aberrant cccDNA, while others integrate into random sites in host DNA. The linear HBV genomes, rather than the circular genomes, appear to be the main precursor of integrated DNA that accumulates during the course of an infection. |
An aberrant linear viral DNA may be formed when positive-sense strand primer translocation fails to occur, so that positive-sense strand synthesis initiates at the 3′-end of the negative-sense strand to create a double-stranded linear (dsl) DNA. The dsl DNA can also circularize to form aberrant cccDNA. A minority of nucleocapsids contains dsl HBV DNA. This dsl DNA cannot serve as a template for further infectious virions after infection of a new cell but can be a substrate for integration into the host genome. Integration is not required for replication but does occur at a low rate. As linearization mandatorily interrupts at least one ORF the integrated DNA cannot code for functional hepadnavirus. Integration of the linear DNA appears to occur via illegitimate recombination. Almost all biopsied hepatocellular carcinoma cells (HCC) have been shown to have integrated viral DNA, often highly rearranged at seemingly random sites. Integrated HBV DNA in HCCs is thought to indicate that HCCs originate by dedifferentiation of normal hepatocytes. However, it is not clear if this has a causative role in most HCC patients.
Biology
All hepadnaviruses show narrow host specificity. In vitro replication of many hepadnaviruses has only been demonstrated following transfection of tissue culture cells by cloned viral DNA, resulting in the production of virus. Replication has been achieved following inoculation of primary hepatocytes with virus-containing serum. Human hepatoma cells transfected to express NTCP support HBV replication.
Hepadnavirus infections in vivo have characteristic features based on the knowledge obtained from the study of avi- and orthohepadnaviruses:
- Avi- and orthohepadnaviruses are markedly hepatotropic, although viral antigens and nucleic acids can also be detected in white blood cells (and in some extra-hepatic sites, e.g. DHBV has been found in pancreas, spleen and kidney). Less is known on the organ tropism for members of the three other genera
- Infection may be transient or persistent, the outcome depending on factors such as host age and dose of inoculum. Persistent infection, which is defined as expression of HBsAg for more than 6 months, is far more common in neonates and in immuno-compromised hosts. Persistent infections are often life-long and accompanied by high levels of virions and subviral particles in the circulating blood.
- Empty subviral particles, composed of excess virus envelope material, are present in much greater numbers than complete virions in most individuals and at most stages of infection.
- Virus replication is generally thought to be non-cytopathic, and different degrees of ongoing liver damage in different individuals are thought to be governed by different degrees of immune-mediated damage to infected hepatocytes.
- In ortho-, but not avihepadnavirus infections, persistent virus infection confers a significantly increased rate of development of primary hepatocellular carcinoma, and a number of direct and indirect mechanisms have been proposed.
Antigenicity
Three principal antigens have been identified for avi- and orthohepadnaviruses, designated surface, core and e antigen. These are abbreviated HBsAg, HBcAg, and HBeAg for the HBV-related antigens, DHBsAg, DHBcAg and DHBeAg for DHBV-related antigens, etc., while the corresponding antibodies are designated anti-HBs, anti-HBc, anti-DHBs, anti-DHBc, etc. HBsAg cross-reacts to a limited extent with the analogous antigen of WHV and ground squirrel hepatitis virus (GSHV) but not with the analogous antigen of DHBsAg. PreS antigens and the HBsAg loop in the S protein bear specific neutralization determinants. SHBs proteins are sufficient to stimulate protective immunity.
Derivation of names
DNA: abbreviation for deoxyribonucleic acid.
Hepa: from Greek hepar, “liver”
Para: from Greek “alongside”
Meta: from Greek “higher”
Herpeto: from Greek “reptile”
Avi: from Latin avis, “bird”
Ortho: from Greek orthos, “straight”
Genus demarcation criteria
The genera are distinguished by the following criteria:
- Nucleotide sequence divergence between complete genomes of viruses belonging to different genera is about 55 % (Table 2. Hepadnaviridae).
- Differences in genome size (about 3.5 for parahepadnaviruses , 3.2 kb for meta- and herpeto- and orthohepadnaviruses, and 3.0 kb for avihepadnaviruses).
- Larger core proteins and no MHBs surface protein for herpeto- and avihepadnaviruses.
- The X protein is essential for orthohepadnaviruses.
- Host range restricted to either teleost fish (meta- and parahepadnaviruses), reptiles and frogs (herpetohepadnaviruses), birds (avihepadnaviruses), and mammals (orthohepadnaviruses).
Table 2. Hepadnaviridae. Nucleotide identity between complete genomes of members of the five genera within the Hepadnaviridae by pairwise comparison of the type species.
| Metahepadnaviruses | Herpetohepadnaviruses | Avihepadnaviruses | Orthohepadnaviruses | |
| Parahepadnaviruses | 62% | 60% | 48% | 52% |
| Metahepadnaviruses | 63% | 55% | 44% | |
| Herpetohepadnaviruses | 54% | 63% | ||
| Avihepadnaviruses | 54% |
Relationships within the family
Phylogenetic relationships between members of the five genera are shown in Figure 4. Hepadnaviridae.
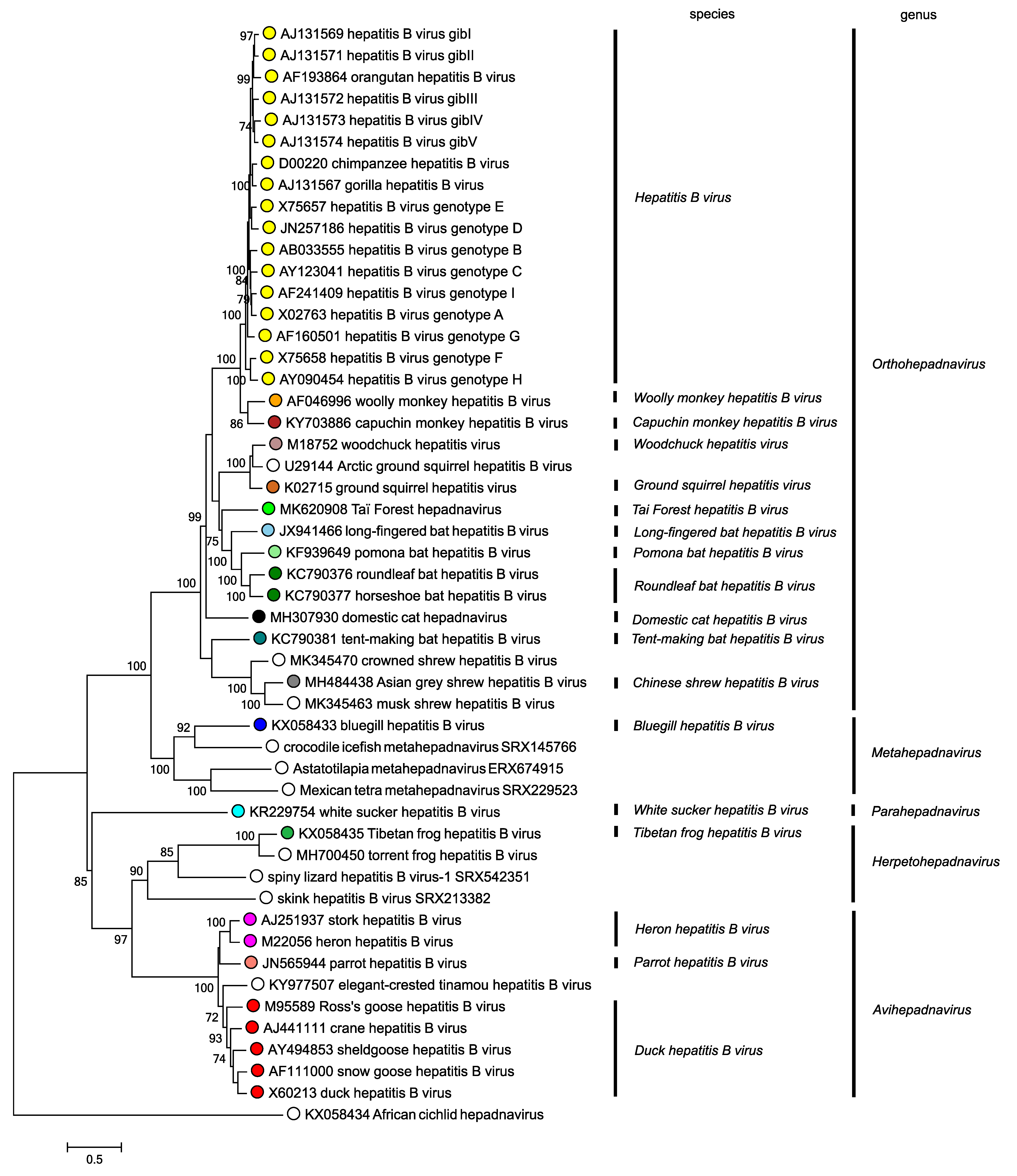 |
| Figure 4. Hepadnaviridae. Phylogenetic tree based on the polymerase genes of 50 selected members of the family Hepadnaviridae. Maximum likelihood trees were produced using the General Time Reversible model with a gamma distribution of variation including invariant sites, and conducted using MEGA7 (Kumar et al., 2016). Numbers indicate where bootstrap support for branches was > 70%. Circles at tips are coloured according to species; unclassified viruses have unfilled circles. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Reverse transcription, as an essential step in replication, is a common feature of members of the families Hepadnaviridae, Nudnaviridae, Caulimoviridae (pararetroviruses), Belpaoviridae, Metaviridae, Pseudoviridae, and Retroviridae (the last five families belonging to the order Ortervirales). Some essential distinguishing features between members of these virus families are given in Table 3. Hepadnaviridae. The virions of pararetroviruses contain dsDNA as nucleic acid, while those of orthoretroviruses, metaviruses, pseudoviruses and belpaoviruses contain ssRNA. Spumaretroviruses contain both ssRNA and dsDNA in extracellular particles. For the pararetroviruses, integration of virus DNA in the host genome is not part of the replication cycle, as it is for retroviruses.
The genomes of hepadna- and retroviruses also contain three major genes, with the same functions, and in the same order (i.e. core, polymerase, and pre-S/S compared to gag, pol, env respectively) although the S-gene of hepadnaviruses is only analogous with those of retroviruses. A fundamental distinction is that, with hepadnaviruses, the form of the genome in extracellular virions is DNA and reverse transcription takes place during the efferent or outgoing arm of the replication cycle, whereas the reverse holds true for retroviruses (with the exception of the spumaviruses, in which some infectious particles appear to contain a DNA genome). Members of the order Ortervirales use tRNAs as primers for synthesis of the DNA negative-sense strand, whereas hepadnaviruses utilize a tyrosine in the polymerase itself. The polymerase protein of hepadnaviruses does not have a protease or integrase function. Many other aspects are distinctly different in members of the Hepadnaviridae and Ortervirales (Krupovic et al., 2018), partly due to the extremely small size of the hepadnaviral genome. The hepadnaviral genome is characterised by considerable overlap of both coding regions and regulatory elements.
Using the TP domain of the P-gene of HBV as a search query in the NCBI public databases [Whole-genome Shotgun Assembly (WGS), Transcriptome Shotgun Assembly (TSA), and Sequence Read Archives (SRA)], a new clade of teleost viruses has been identified. These share many features with members of the Hepadnaviridae and are therefore considered a sister family to this family (Lauber et al., 2017). They were designated as nackednaviruses, a name given due to their absence of an envelope, which is a fundamental difference to hepadnaviruses. The provisional taxon designation Nudnaviridae is proposed here for this family.
Table 3. Hepadnaviridae. Features distinguishing the members of Hepadnaviridae, the provisional virus family Nudnaviridae, and families within the order Ortervirales
| Ortervirales | ||||||||
Hepadna-viridae |
Nudna- viridae |
Caulimo-viridae | Retroviridae |
Belpao-viridae |
Meta-viridae |
Pseudo-viridae | ||
| Orthoretro-virinae | Spumaretro-virinae | |||||||
| Envelope | Yes | No | No | Yes | Yes | Yes/No | Yes | No |
| Genome in virion | DNA | DNA | DNA | RNA | RNA & DNA | RNA | RNA | RNA |
| Genomic size | 3.0-3.4 kb | 2.7-3.1 kb | 6.0-8.0 kb | 8.0-10.0 kb | 10.5-11.7 kb | 6-8 kb | 4-10 kb | 5-9 kb |
| Viral encoded protease / integrase | No/No | No/No | Yes/No | Yes/Yes | Yes/Yes | Yes/Yes | Yes/Yes | Yes/Yes |
| Integration into the host genome for replication | No | No | No | Yes | Yes | Yes | Yes | Yes |
| Host | Vertebrates | Teleost fish | Plants, angiosperms | Vertebrates | Mammals | Eukaryotes | Eukaryotes | Eukaryotes |

