Family: Hepeviridae
Michael A. Purdy, Jan Felix Drexler, Xiang-Jin Meng, Heléne Norder, Hiroaki Okamoto, Wim H. M. Van der Poel, Gábor Reuter, William M. de Souza, Rainer G. Ulrich and Donald B. Smith
The citation for this ICTV Report chapter is the summary published as Purdy et al., (2022):
ICTV Virus Taxonomy Profile: Hepeviridae 2022, Journal of General Virology, (2022) 103:001778.
Corresponding author: Donald B. Smith ([email protected])
Edited by: Nick J. Knowles and Peter Simmonds
Posted: November 2017, updated June 2022
PDF: ICTV_Hepeviridae.pdf (2017 version)
Summary
The family Hepeviridae includes enterically-transmitted, small, quasi-enveloped viruses with positive-sense RNA genomes (Table 1.Hepeviridae). Members of the family are assigned to two subfamilies, five genera and ten species. Members of the subfamily Parahepevirinae infect trout and salmon, and members of the subfamily Orthohepevirinae infect mammals and birds. The species Paslahepevirus balayani and Rocahepevirus ratti include hepeviruses which can cause a self-limited acute hepatitis of zoonotic origin in humans and several mammalian species, but may become chronic in immunocompromised patients. Extrahepatic manifestations of neurological sequelae such as Guillain–Barré syndrome and neuralgic amyotrophy, renal and pancreatic diseases such as glomerulonephritis and pancreatitis have been described in humans. The species Avihepevirus magniiecur includes avian HEV that causes hepatitis-splenomegaly syndrome in chickens. Members of the species Piscihepevirus heenan infect salmonid fishes, but are not known to cause disease.
Table 1.Hepeviridae. Characteristics of members of the family Hepeviridae.
| Characteristic | Description |
| Example | hepatitis E virus Burma (M73218), species Paslahepevirus balayani, genus Paslahepevirus |
| Virion | Quasi-enveloped, 27–34 nm diameter with a single capsid protein |
| Genome | 6.4-7.2 kb capped positive-sense monopartite RNA containing 3 open reading frames |
| Replication | Occurs in association with the host endoplasmic reticulum. |
| Translation | From genomic (ORF1) and subgenomic (ORF2 and ORF3) capped RNA |
| Host range | Mammals (Chirohepevirus, Paslahepevirus, Rocahepevirus), birds (Avihepevirus) and salmonid fish (Piscihepevirus) |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Kitrinoviricota, class Alsuviricetes, order Hepelivirales; two subfamilies, five genera and ten species |
Virion
Morphology
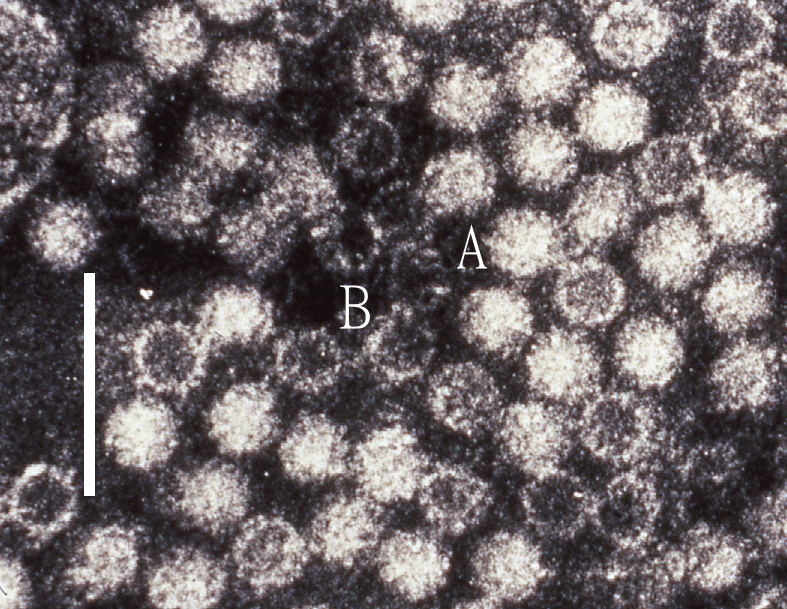
Virions of hepeviruses are icosahedral, quasi-enveloped, spherical particles with a diameter of approximately 27–34 nm (Figure 1.Hepeviridae). Spikes and indentations can be seen on electron micrographs of virions (Bradley 1990). The capsid is formed from capsomeres consisting of homodimers of a single capsid protein, forming the virus shell. Each capsid protein contains three linear domains forming distinct structural elements: S (the continuous capsid), P1 (three-fold protrusions), and P2 (two-fold spikes). Neutralizing epitopes have been found in the P2 domain. Each domain contains a putative polysaccharide-binding site that may interact with cellular receptors. Native T=3 capsids contain flat dimers, with less curvature than those of T=1 virus-like particles (Figure 2.Hepeviridae) (Mori and Matsuura 2011). Virions released into blood have a lower density in sucrose than do virions from faeces, this difference being associated with the incorporation of lipids and the protein encoded by ORF3 (Takahashi et al., 2010). Virions of avian HEV (Figure 3.Hepeviridae) revealed by negative staining electron microscopy (EM) of bile samples from chickens with hepatitis-splenomegaly syndrome are un-enveloped and are similar in size and morphology to those of members of the species Orthohepevirus balayani (Haqshenas et al., 2001).
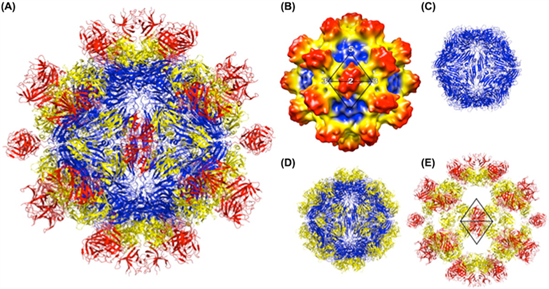 |
| Figure 2.Hepeviridae. Structure of the hepatitis E virus-like particle (VLP) (T=1). (A) Crystal structure of hepatitis E virus VLP. The three domains, S, P1 and P2 are colored blue, yellow and red, respectively. The VLP is positioned in a standard orientation with the three 2-fold icosahedral symmetry axes aligned along the vertical, horizontal, and viewing directions, respectively. (B) Cryo-EM reconstruction at 14 Å resolution. The surface is coloured by radial depth cue from blue, yellow, to red. (C) Hepatitis E virus VLP with only the S domain. (D) VLP with S and P1 domains. (E) VLP with P1 and P2 domains (from (Guu et al., 2009) with permission). |
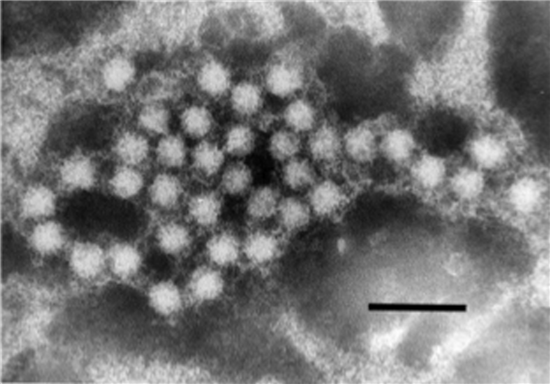 |
| Figure 3.Hepeviridae. Electron micrograph of 30–35 nm diameter particles of avian hepatitis E virus. Virus particles were detected from a bile sample of a chicken with hepatitis-splenomegaly syndrome. Bar = 100 nm (from (Haqshenas et al., 2001) with permission from the Microbiology Society). |
Physicochemical and physical properties
Non-enveloped virions of members of the subfamily Orthohepevirinae have buoyant densities of 1.35 to 1.40 g cm−3 in CsCl and 1.29 g cm−3 in glycerol and potassium tartrate gradients. The virion S20,w is 183S. Virions are sensitive to low-temperature storage (between −70°C and +8°C) and iodinated disinfectants (Bradley 1990). The virion of HEV is more heat-labile than that of hepatitis A virus: HEV was about 50% inactivated at 56°C, but 1% infectivity remained after 60 min at this temperature; no infectivity was detected after heating at 66°C or 70°C for 1 h (Emerson et al., 2005). Liver suspensions containing avian HEV remained infectious after treatment with chloroform and ether but lost infectivity after incubating at 56°C for 1 h or 37°C for 6 h. Viral infectivity in liver suspensions was reduced 1000-fold after treatment with 0.05% Tween-20, 0.1% NP40 and 0.05% formaldehyde (Meng et al., 2006). Properties of members of the subfamily Parahepevirinae have not been investigated.
Nucleic acid
The genome of members of the family Hepeviridae is a linear, positive-sense, single-stranded RNA molecule of approximately 6.6–7.2 kb, with a m7G cap structure at the 5′-end and a poly(A) tail at the 3′-end.
Proteins
Virions are constructed from a major capsid protein (CP) encoded by the second open reading frame (ORF2). The CP binds to surface heparan sulfate proteoglycans (HSPGs) on liver cells (Kalia et al., 2009); there is evidence that the form of CP present on virions is C-terminally truncated (Yin et al., 2018, Ankavay et al., 2019, Nishiyama et al., 2021). A small immunoreactive protein (112–114 amino acids, 12.5 kDa) encoded by the third ORF (ORF3) is associated with virion particles in serum and has been shown to exhibit multiple functions associated with virion morphogenesis, egress and viral pathogenesis (Tanggis et al., 2018). The ORF3 polypeptide has been shown to share several structural features with class I viroporins (Ding et al., 2017).
Lipids
Although HEV is shed in faeces as a non-enveloped virus there is evidence that, like hepatitis A virus (Picornaviridae), HEV can hijack host membranes on assembly and exit. Possession of a host-derived envelope may allow the virus to circulate in a patient’s blood escaping detection by neutralizing antibodies (Yin et al., 2016).
Carbohydrates
Evidence for glycosylation of the major CP has been reported following its expression in mammalian cells (Jameel et al., 1996). The CP sequence contains three potential sites for N-linked glycosylation and a signal peptide sequence at its amino-terminus (Zafrullah et al., 1999). Amino acid exchanges within HEV CP glycosylation sites prevent the formation of infectious virus particles, although the lethal effect is due to altered protein structure rather than elimination of glycosylation (Graff et al., 2008).
Genome organization and replication
The RNA genome of members of the family Hepeviridae is organized into three ORFs, with the non-structural proteins encoded toward the 5′-end of the genome and the structural protein(s) toward the 3′-end. An additional ORF (ORF4) overlapping the start of ORF1 is found in strains belonging to the species Orthohepevirus ratti (Johne et al., 2010a, Johne et al., 2014a), although in vitro studies have failed to detect a protein product, and lack of expression or ORF4 did not affect virus growth in cell culture (Tanggis et al., 2018). Capped genomic RNA of an Avihepevirus magniiecur isolate has been shown to be infectious for chickens (Huang et al., 2005a) and that of Paslahepevirus balayani for pigs, rhesus monkeys and chimpanzees (Panda et al., 2000, Emerson et al., 2001, Huang et al., 2005b). The 5′-non-coding region (NCR) is up to 34 nt in length among members of the species Paslahepevirus balayani but is 100 nt in cutthroat trout virus (Piscihepevirus heenan). A methylguanine cap structure has been identified at the 5′ end of the hepevirus genome and plays a role in the initiation of virus replication (Ahmad et al., 2011). The 3′-NCR of hepeviruses contains a cis-reactive element (Emerson et al., 2001) as does a central region of ORF2 (Emerson et al., 2013). The 3′-end of the genome is also polyadenylated.
In all members of the family Hepeviridae, ORF1 encodes a non-structural polyprotein, followed by ORF2 that encodes the CP, and an overlapping reading frame, ORF3 which encodes a small phosphoprotein of 112–114 aa with a multifunctional carboxy-terminal region (Figure 4.Hepeviridae). A bicistronic subgenomic mRNA encoding both CP and ORF3 protein has been identified (Graff et al., 2006). Isolates have been described from chronically infected individuals that contain an insertion of host-derived sequences such as human ribosomal protein S17 within the ORF1 hypervariable region that confers a growth advantage in cultured hepatoma cells (Shukla et al., 2011, Nguyen et al., 2012, Johne et al., 2014b).
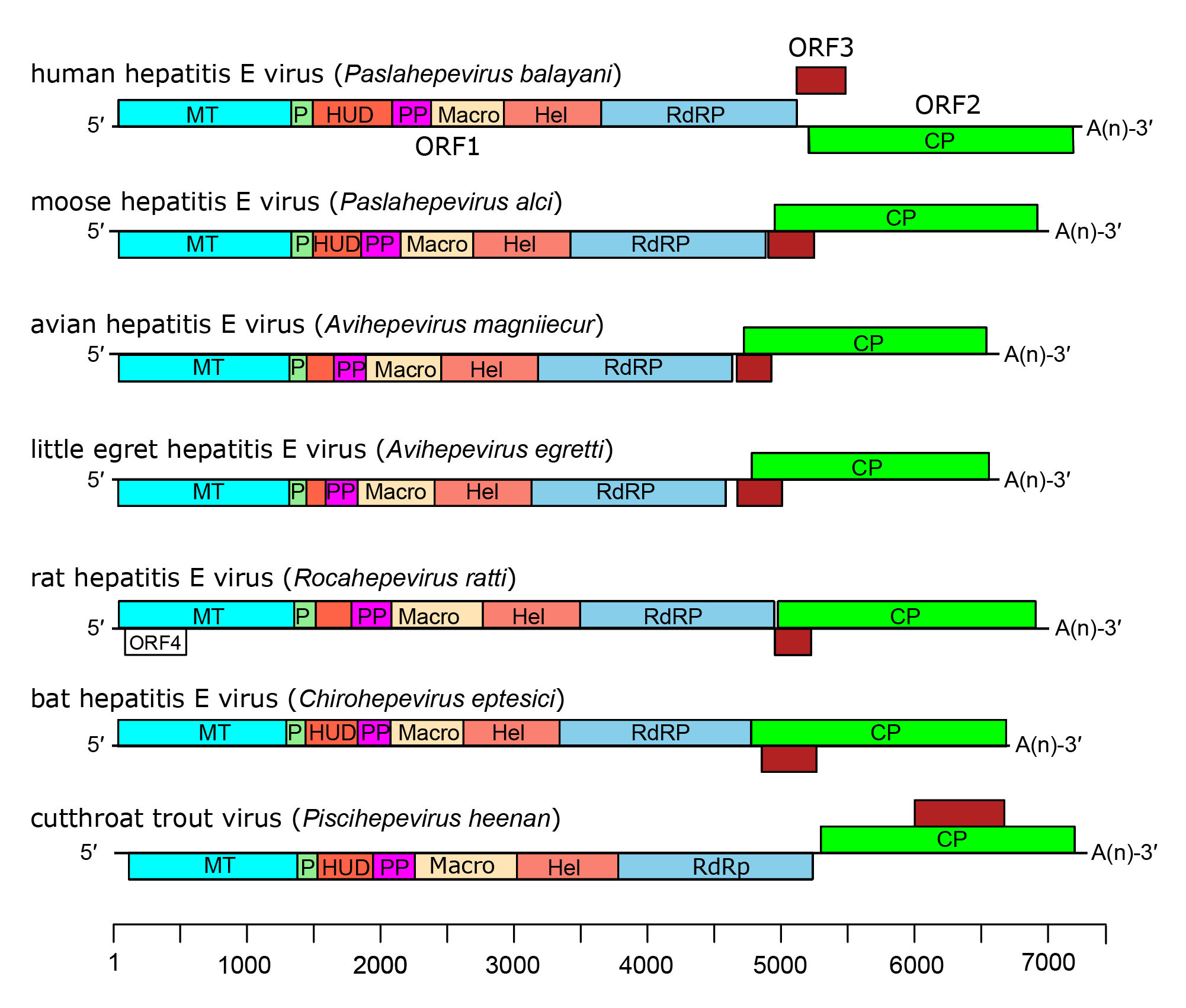 |
| Figure 4.Hepeviridae. Genome organization of members of the family Hepeviridae. ORFs are shown as boxes between short 5′-and 3′-non-coding regions. ORF1 encodes non-structural proteins including putative functional domains MT, methyltransferase; P, a putative papain-like cysteine protease; HUD, hepevirus unique domain also called the Z domain (Kelly et al., 2016); PP, a hypervariable polyproline region that is dispensable for virus infectivity; Hel, helicase; RdRP, RNA-directed RNA polymerase (Koonin et al., 1992, Kelly et al., 2016). ORF2 (green) encodes capsid protein and ORF3 (brown) encodes a small phosphoprotein with a multi-functional C-terminal region. ORF2 and ORF3 overlap each other but neither overlaps ORF1. For members of the species Orthohepevirus ratti a conserved fourth ORF overlaps ORF1, putatively encoding a short protein of unknown function. |
Non-structural proteins encoded by the first major ORF (ORF1) show some similarity with the “alpha-like supergroup” of viruses, with domains potentially encoding a methyltransferase, a putative papain-like cysteine protease, a macro domain, an RNA helicase and an RNA-directed RNA polymerase (RdRP) (Cao and Meng 2012). Some of these predicted enzymatic properties have been confirmed experimentally (Karpe and Lole 2010, Parvez 2015, Mahilkar et al., 2016). A hypervariable region lies between the protease and helicase domains. The hypervariable region contains two subregions; the amino-terminal half of this region consists of a variable length polypeptide with host species-specific sequence conservation, which is unique to members of the family Hepeviridae (Kelly et al., 2016) and may be responsible for host specificity (Lara et al., 2014). The carboxyl-terminal half consists of an intrinsically-disordered polypeptide with a high frequency of proline residues (Purdy 2012, Kelly et al., 2016). The translational and post-translational processing of the non-structural polyprotein remains unresolved. In particular, it remains unclear whether the non-structural polyprotein functions as a single protein with multiple functional domains or whether it is proteolytically cleaved into smaller proteins with distinct enzymatic activities. CP, encoded by ORF2, can be glycosylated, although these modified forms are not present in virions (Yin et al., 2018, Ankavay et al., 2019). There is also evidence that a short form of CP, resulting from use of an alternate initiation site or produced through proteolytic cleavage, is present in infectious virions (Yin et al., 2018, Ankavay et al., 2019, Nishiyama et al., 2021).
Replication of hepeviruses is not well understood. The viral RdRP associates with the host endoplasmic reticulum (ER) through ORF1 amino acid residues 1189−1391 encoding a predicted transmembrane domain to begin replicating the viral genome. It appears that replication involves temporal separation and alternating cycles of positive- and negative-sense RNAs to produce capsid, ORF3 protein, ORF1 polypeptide, and new genomes, resulting in the generation of progeny virions (Varma et al., 2011).
Biology
Hepatitis E virus is the only hepatitis virus of humans that is transmitted zoonotically (Rasche et al., 2019). Hepevirus infection occurs by the faecal-oral route for members of the species Paslahepevirus balayani and Avihepevirus magniiecur, and is usually associated with a self-resolving acute infection of the liver. More recently, chronic hepatitis E has become a significant clinical problem in immunosuppressed individuals especially in solid organ transplant recipients. Transmission can occur through contaminated water, consumption of raw/undercooked meats or faeces from infected animals (e.g. pigs, roe deer, wild boar), and rarely through blood transfusion, transplantation or xenotransplantation. Hepatitis E virus can be vertically transmitted from mother to foetus. Rat hepatitis E virus (species Rocahepevirus ratti) can be a zoonotic pathogen for humans (Sridhar et al., 2018, Andonov et al., 2019, Reuter et al., 2020, Sridhar et al., 2021, Rivero-Juarez et al., 2022). Little is known about the mode of transmission or pathologenicity for members of other species in the family.
Antigenicity
Infected individuals typically develop antibodies directed against the CP. Cross-reactivity has been demonstrated between the capsid proteins of viruses of the species Paslahepevirus balayani and Avihepevirus magniiecur (Haqshenas et al., 2002), and antibodies raised against the CP from dromedary camel-associated HEV, expressed in baculovirus expression system, exhibited antigenic cross-reactivity against several genotypes of virus strains belonging to the species Paslahepevirus balayani as well as rat and ferret HEVs belonging to the species Rocahepevirus ratti (Zhou et al., 2015). A monoclonal antibody raised against HEV-3 CP confirmed a cross-reactivity to recombinant and cell free-produced CP derivatives of several genotypes of Paslahepevirus balayani, Orthohepevirus ratti (genotype C1), bat and common vole hepeviruses (Kubickova et al., 2021). Given the diversity of hosts from which hepeviruses have now been described, reports of serological reactivity to HEV antigens in novel host species are difficult to interpret without corresponding virus sequence information.
Vaccines capable of protecting against human infection with HEV have been produced using portions of the capsid protein (Purdy et al., 1993, Tsarev et al., 1997) and tested in Nepal (Shrestha et al., 2007), and China (Zhu et al., 2010). Xiamen Biotech has a vaccine that is licenced in China, but that is not currently licensed for use in other countries. A vaccine comprising part of the Avihepevirus magniiecur capsid protein has been shown to protect chickens against infection (Guo et al., 2007).
Derivation of names
Avihepevirus: from avian and hepatitis E virus
Avihepevirus magniiecur - from the Latin for big liver - magni iecur
Chirohepevirus: from Chiroptera, the order to which bats belong, and hepatitis E virus
Hepeviridae: from hepatitis E virus.
Orthohepevirinae: from the Greek orthos, meaning straight, and hepatitis E virus
Parahepevirinae: from the Greek para, meaning by the side of, and hepatitis E virus
Paslahepevirus: from Primates, Artiodactyla, Scandentia, Lagomorpha, the taxa of known hosts of members of the genus
Paslahepevirus balayani: refers to the first hepatitis E virus paper (Balayan et al., 1983)
Piscihepevirus: from the Latin piscis, meaning fish, and hepatitis E virus
Piscihepevirus heenan: from Heenan Lake, the geographic origin of the first member of the species
Rocahepevirus: from Rodentia & Carnivora, the known hosts of members of the genus
Species epithets are Latinised versions of a host genus name (Table 2.Hepeviridae), except for Paslahepevirus balayani which refers to the first paper on human HEV (Balayan et al., 1983), Avihepevirus magniiecur, derived from the Latin for big liver (magni iecur) and Piscihepevirus heenan, which refers to Heenan Lake where strains of this species were first discovered.
Table 2.Hepeviridae Previous and current species names
| Previous species name | Member viruses | Current species name |
| Orthohepevirus A | HEV of humans, pigs, deer, wild boar, rabbits, camels etc. | Paslahepevirus balayani |
| Unassigned | HEV of moose | Paslahepevirus alci |
| Orthohepevirus B | avian HEV (chickens) | Avihepevirus magniiecur |
| Unassigned | avian HEV (Little egret) | Avihepevirus egretti |
| Orthohepevirus C | HEV of rats, ferrets, minks, field mice | Rocahepevirus ratti |
| Unassigned | HEV of voles | Rocahepevirus eothenomi |
| Orthohepevirus D | HEV of Eptesicus and Myotis bats | Chirohepevirus eptesici |
| Unassigned | HEV of Desmodus bats | Chirohepevirus desmodi |
| Unassigned | HEV of Rhinolophus bats | Chirohepevirus rhinolophi |
| Piscihepevirus A | HEV of trout and salmon | Piscihepevirus heenan |
Subfamily demarcation criteria
Members of different subfamilies differ in host range with members of the subfamily Parahepevirinae only known from fish, while members of the subfamily Orthohepevirinae have been detected in a wide range of mammals and birds (Spahr et al., 2018). Members of the two subfamilies also differ in phylogenetic relationships observed for comparisons between three conserved regions within ORF1. These relationships are mirrored in sequence distance between representatives of each subfamily with inter-subfamily amino acid p-distances greater than 0.6 compared to intra-subfamily p-distances of less than 0.5 (Smith et al., 2014, Smith et al., 2015). Members of the same subfamily all share similar genome organisation: in members of the subfamily Orthohepevirinae ORF3 overlaps the 5′-end of ORF2 while in piscihepeviruses the overlap is more central (Batts et al., 2011).
Relationships within the family
The family includes two subfamilies whose members are phylogenetically distinct; the subfamily Orthohepevirinae includes four genera that are phylogenetically distinct and have different host ranges (Figure 5.Hepeviridae). Only one species has been described in the subfamily Parahepevirinae.
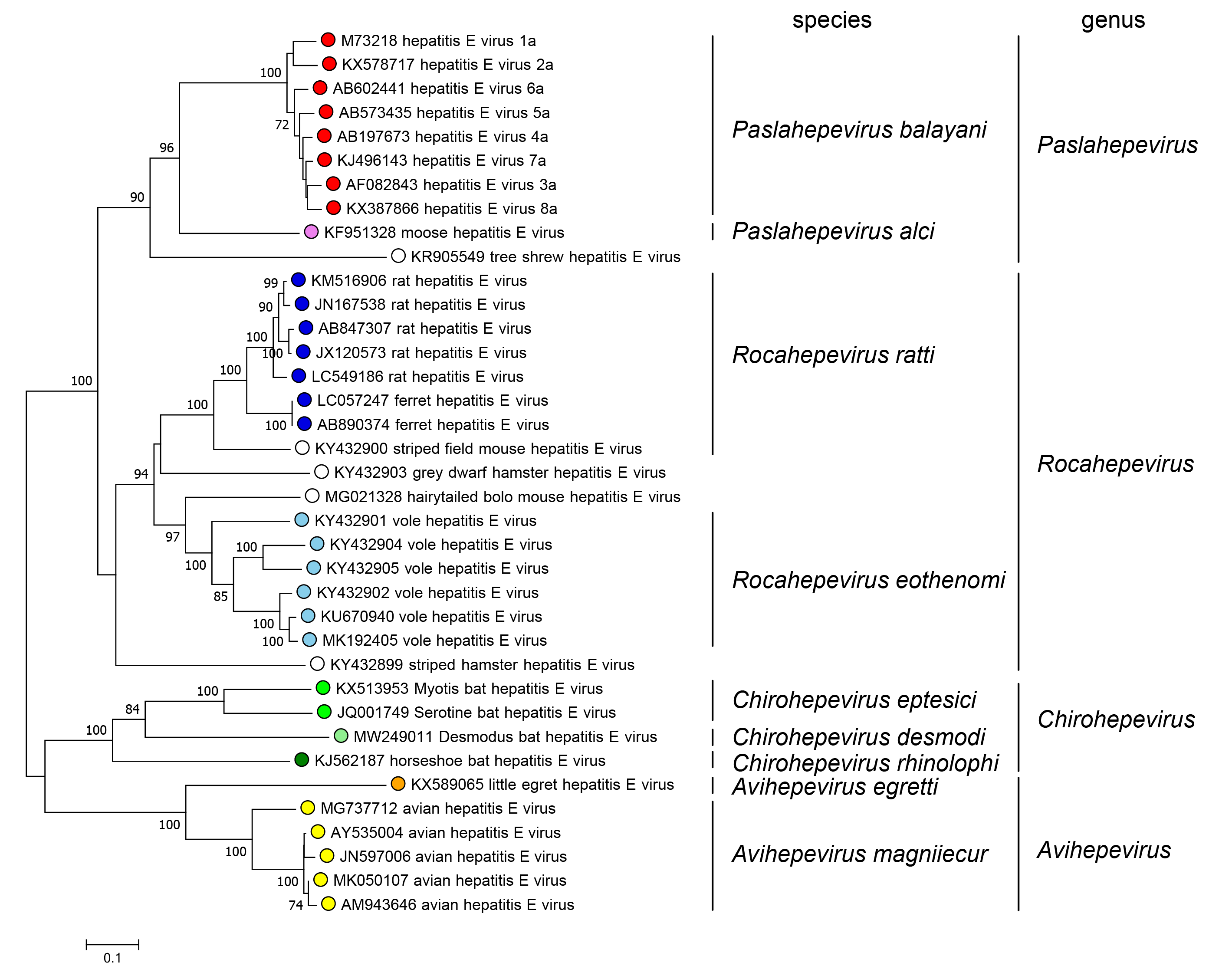 |
| Figure 5.Hepeviridae. Phylogenetic tree of members of the subfamily Orthohepevirinae. An alignment of the ORF1 polyprotein residues 1–450, corresponding to the methyltransferase domain generated using MUSCLE (Edgar 2004) was analysed in MEGA7 (Kumar et al., 2016) using the JTT method and a gamma distribution of rate variation between sites. Numbers indicate bootstrap support for nodes where this was over 70%. Dots at branch tips are coloured by species; unfilled dots are unclassified. |
Relationships with other taxa
Hepeviruses are similar to members of the family Caliciviridae based on the superficial structural morphology as revealed by EM, and its genome organization (Bradley 1990). However, members of the two families show little detectable sequence similarity and the cap structure at the 5′-end of the hepevirus genome is absent in caliciviruses. Hepeviruses show highest, but limited, amino acid sequence similarity in its replicative enzymes with rubella virus (Rubivirus rubellae, family Matonaviridae), alphaviruses of the family Togaviridae and with plant furoviruses. The capping enzyme, helicase and replicase of hepeviruses have properties very similar to those of viruses within the “alphavirus-like supergroup” (Kelly et al., 2016).
Related, unclassified viruses
| Virus | Source | Accession number |
| bivalve hepelivirus G | bivalves | KX158876 |
| Dongbei arctic lamprey hepevirus | Lethenteron camtschaticum | MG600001 |
| Hepelivirus* | sewage | JQ898340 |
| Hepelivirus* | chimpanzee | KM407530 |
| Rana hepevirus | agile frog (Rana dalmatina) | MH330682 |
| Wenling fish hepevirus | fish | MG600002 |
| Wenling moray eel hepevirus | Gymnothorax reticularis | MG600005 |
| Wenling samurai squirrelfish hepevirus | Sargocentron ittodai | MG600003 |
| Wenling thamnaconus septentrionalis hepevirus | Thamnaconus septentrionalis | MG600004 |
| Wenling thamnaconus striatus hepevirus | Thamnaconus striatus | MG600006 |
Virus names and virus abbreviations are not official ICTV designations.
* partial sequence
The taxonomic position of these novel viruses, variously described as hepeviruses and hepeliviruses is unclear (Ng et al., 2012, Zhou et al., 2014, Rosani and Gerdol 2017, Reuter et al., 2018, Shi et al., 2018). While they share a similar genome organisation to members of the family Hepeviridae, they are phylogenetically distant, and may represent members of novel families within the order Hepelivirales, rather than additional taxa in the family Hepeviridae.