Family: Nanoviridae
John E. Thomas, Bruno Gronenborn, Robert M. Harding, Bikash Mandal, Ioana Grigoras, John W. Randles, Yoshitaka Sano, Tania Timchenko, H. Josef Vetten, Hsin-Hung Yeh and Heiko Ziebell
The citation for this ICTV Report chapter is the summary published as Thomas et al. (2021)
ICTV Virus Taxonomy Profile: Nanoviridae, Journal of General Virology, 102 (3): 001544
Corresponding author: John E. Thomas ([email protected])
Edited by: F. Murilo Zerbini and Sead Sabanadzovic
Posted: November 2020, updated January 2025
PDF: ICTV_Nanoviridae.pdf
Summary
The family Nanoviridae comprises plant viruses with a multipartite, circular, single-stranded DNA genome (6 or 8 components), allocated to two genera, Babuvirus and Nanovirus (Table 1 Nanoviridae). The small, isometric virions (17–19 nm diameter) each contain a single genome component, and are transmitted by aphids in a circulative, non-propagative persistent manner.
Table 1 Nanoviridae. Characteristics of members of the family Nanoviridae
| Characteristic | Description |
| Example | subterranean clover stunt virus [isolate Myall Vale 2534B] (DNA-R: MK035728; DNA-S: MK035729; DNA-C: MK035730; DNA-M: MK035731; DNA-N: MK035732; DNA-U1: MK035733; DNA-U2: MK035734; DNA-U4: MK035735), species Nanovirus trifolii, genus Nanovirus |
| Virion | 17–19 nm isometric, containing a single type of capsid protein |
| Genome | Multipartite, circular single-stranded DNA, comprising 6 (each 1.0–1.1 kb; Babuvirus) or 8 (each 0.9–1.0 kb; Nanovirus) components |
| Replication | Nuclear, by rolling-circle replication using host DNA polymerase |
| Translation | Via dsDNA intermediates with the aid of host DNA and RNA polymerases |
| Host range | Eudicots, mainly Fabaceae (genus Nanovirus); monocotyledons, order Zingiberales (genus Babuvirus); transmitted by specific aphid vectors |
| Taxonomy | Realm Monodnaviria, kingdom Shotokuvirae, phylum Cressdnaviricota, class Arfiviricetes, order Mulpavirales; two genera including 15 species |
Virion
Morphology
Virions are 17–19 nm in diameter, and presumably of an icosahedral T=1 symmetry structure made from 60 capsid protein subunits (Chu and Helms 1988, Wu and Su 1990). They are not enveloped. The virions can be angular or hexagonal in outline when viewed along their 3-fold axis of symmetry (Figure 1 Nanoviridae).
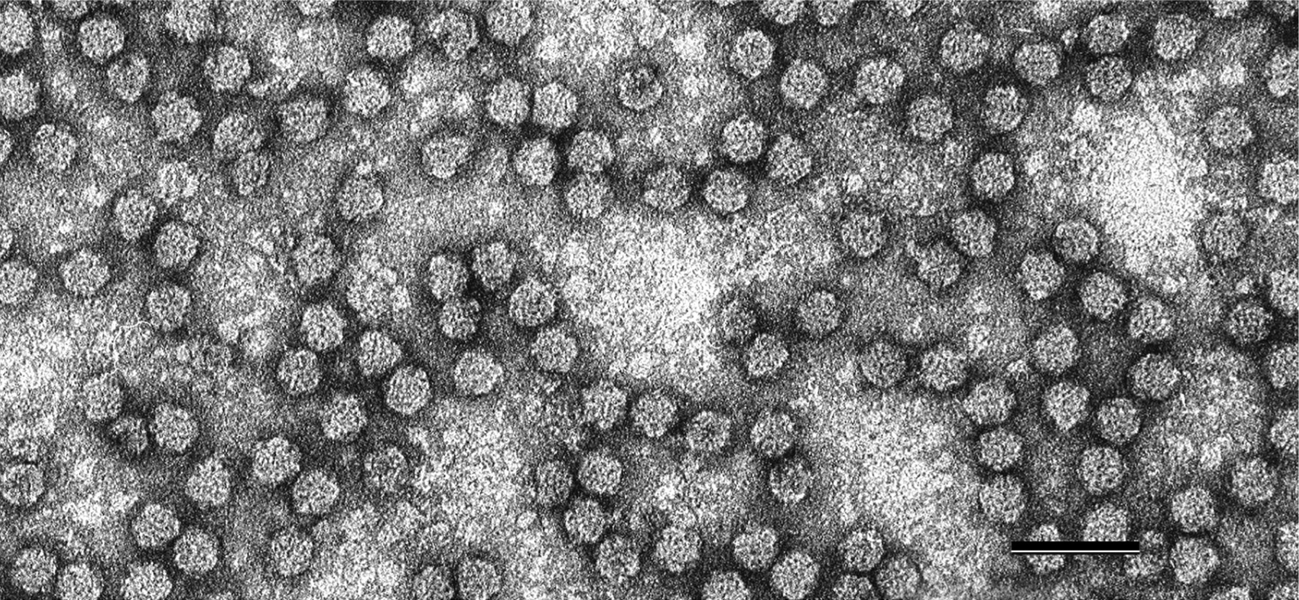 |
| Figure 1 Nanoviridae. Negative-contrast electron micrograph of particles of faba bean necrotic yellows virus. The bar represents 50 nm. (Courtesy of L. Katul and D.-E. Lesemann.) |
Physicochemical and physical properties
Virions sediment as a single component in sucrose rate-zonal density gradients, and the sedimentation coefficient of banana bunchy top virus (BBTV) virions is 46 S. The buoyant density of virions is about 1.24 to 1.30 g cm−3 in Cs2SO4, and 1.34 g cm−3 in CsCl (Chu and Helms 1988, Thomas and Dietzgen 1991). The particle weight of subterranean clover stunt virus (SCSV) is approximately 1.6×106. SCSV has an A260 nm/A280 nm ratio of 1.35 and an extinction coefficient (corrected for light scattering) of 3.6 at A260 nm (Chu and Helms 1988).
Nucleic acid
Individual nanovirids are consistently associated with either six or eight separately encapsidated positive-sense, single-stranded (ss), circular (c) genomic DNAs, ranging from 917 to 1114 nucleotides (Grigoras et al., 2014, Grigoras et al., 2009, Gronenborn 2004, Stainton et al., 2015, Gronenborn and Vetten 2020). Additionally, varying numbers of separately encapsidated satellite-like sscDNAs (alphasatellites), of about 1000–1100 nt, are associated with many nanovirid isolates (Briddon et al., 2018).
Proteins
Virions have a single type of capsid protein (CP) of about 19 kDa, and no other proteins have been found associated with virions (Chu and Helms 1988, Thomas and Dietzgen 1991).
Lipids
Not present
Carbohydrates
Not present
Genome organization and replication
Babuviruses and nanoviruses have six and eight DNAs, respectively, that comprise the core genome (Figure 2 Nanoviridae). The nanovirus DNA components range from 917 to 1046 nt, while those of the babuviruses are slightly larger (1013–1114 nt). All nanovirid DNAs have a similar structural organization, including conserved inverted repeat sequences potentially forming a stem-loop structure that are the origin of replication and part of the common region-stem loop [CR-SL], and a second common region named CR-M (for babuviruses) or CR-II (for nanoviruses) (Gronenborn et al., 2011, Hu and Yeh 2011) (Figure 2 Nanoviridae). The additional satellite-like DNAs contain different stem-loop sequences.
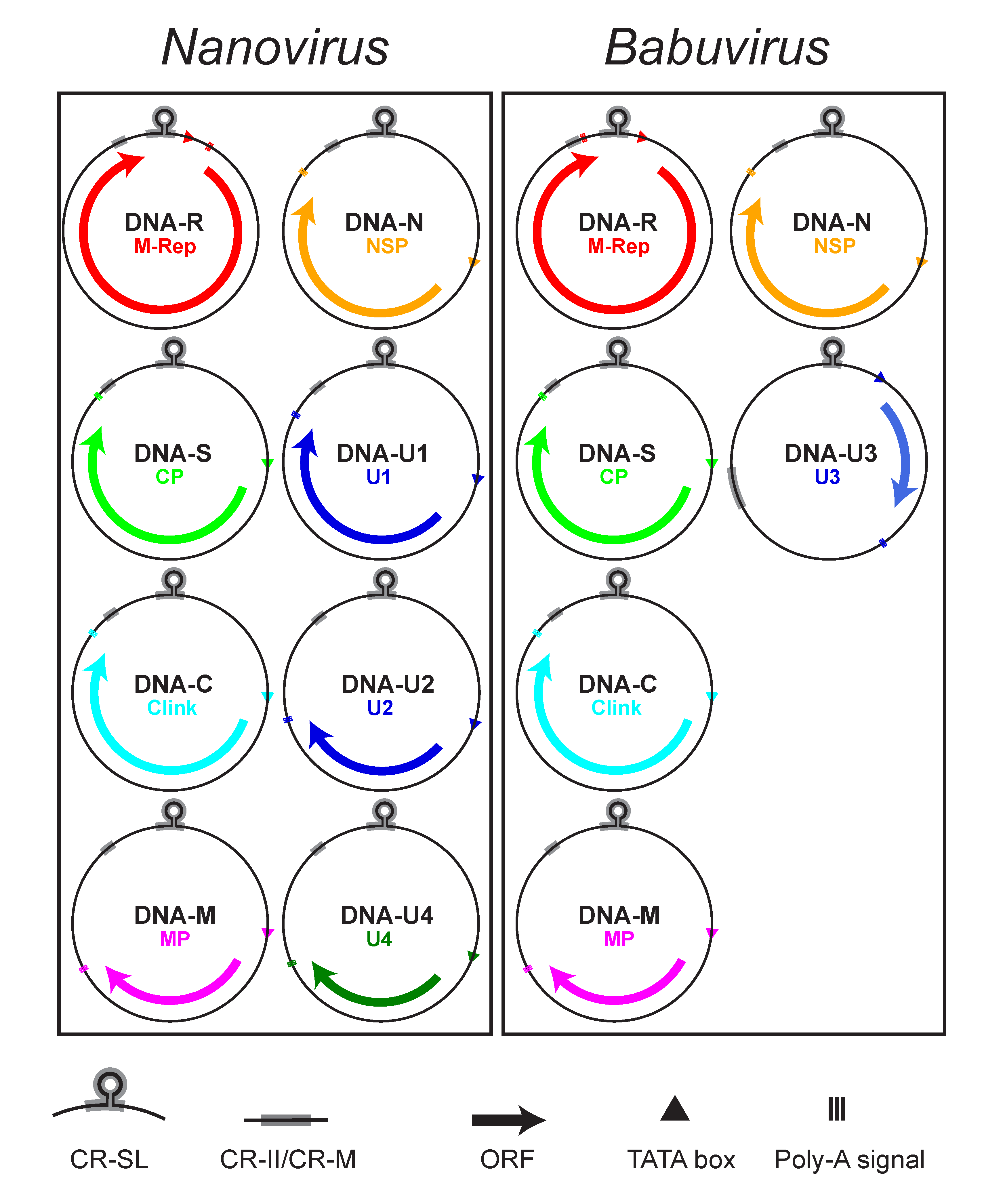 |
| Figure 2 Nanoviridae. Diagram illustrating the genomic organization of viruses of the family Nanoviridae. Each DNA circle (0.9–1.1 kb) is labelled with its name and that of the encoded protein. Arrows refer to the location and approximate size of the ORFs and the direction of mRNA transcription from the dsDNA intermediate. The positions of the common stem-loop region (CR-SL) and the second common region (CR-II/CR-M) are indicated. |
In addition, at least 5–7 non-structural proteins are encoded by the mRNA(s) transcribed from nanovirid dsDNA intermediates (Table 2 Nanoviridae). All but one of the nanovirid DNAs encode a single protein (Beetham et al., 1999, Katul et al., 1998). A second virion-sense ORF, completely nested within the M-Rep-encoding ORF and encoding a putative 5.2 kDa protein of unknown function (U5), has been identified and shown to be transcribed from banana bunchy top virus (BBTV) DNA-R (Beetham et al., 1997). An internal ORF of similar size (5.1–5.2 kDa) and location is present in cardamom bushy dwarf virus (CBDV) but its amino acid sequence is unrelated to that of BBTV. Such an ORF is not present in abaca bunchy top virus (ABTV) or in nanoviruses.
Table 2 Nanoviridae. Proteins encoded by members of the genera Nanovirus and Babuvirus.
| Proteina | Protein mass (kDa) | Encoded by DNA component | Nanoviruses | Babuviruses | Function(s) |
| M-Rep | 33.1–33.7 | DNA-R | + | + | Replication initiator protein for all genomic DNAs |
| CP | 18.7–19.6 | DNA-S | + | + | Structural protein, virion formation (encapsidation) |
| Clink | 18.5–19.9 | DNA-C | + | + | Cell cycle regulation |
| MP | 12.7–13.7 | DNA-M | + | + | Cell-to-cell movement, suppressor of RNA silencing |
| NSP | 17.3–17.7 | DNA-N | + | + | Presumed nuclear shuttle protein (by analogy to geminiviruses), helper factor for aphid transmission |
| U1 | 16.9–18.0 | DNA-U1 | + | − | Unknown |
| U2 | 14.2–15.4 | DNA-U2 | + | − | Unknown |
| U3 | 10.3 | DNA-U3 | − | + | Unknown |
| U4 | 10 or 12.5 | DNA-U4 | + | − | Unknown |
Viruses in the two genera share a set of five homologous DNA components, referred to as DNA-R (encoding the Master Replication Initiator Protein, M-Rep), DNA-S (encoding the capsid protein, CP), DNA-C (encoding the Clink protein), DNA-M (encoding the movement protein, MP) and DNA-N (encoding the nuclear shuttle protein, NSP) (Table 2. Nanoviridae, Figure 2. Nanoviridae). DNAs encoding proteins of unknown function have been identified from nanoviruses (DNA-U1, DNA-U2 and DNA-U4) and babuviruses (DNA-U3, translatable in some BBTV isolates only) (Gronenborn et al., 2011, Vetten 2009).
All nanovirid DNAs contain a major virion sense ORF (Vetten 2009). Uniquely, two mRNAs (putatively encoding M-Rep and U5 proteins) are transcribed from the BBTV DNA-R (Beetham et al., 1997), although a translated U5 protein has yet to be identified in BBTV-infected plants. Each coding region is preceded by a promoter sequence with a TATA box and is followed by a polyadenylation signal (Figure 2. Nanoviridae), except for the nanovirus DNA-R in which the polyadenylation signal is located between the TATA box and the ORF. This leads to transcription of the replication origin and synthesis of a terminally redundant mRNA that is capable of folding into extended secondary structures, and may serve to regulate the expression of the encoded master replication initiator (M-Rep) protein (Grigoras et al., 2008).
The replication of nanovirids is thought to occur in the nucleus by a rolling-circle replication mechanism, through transcriptionally active dsDNA intermediates (Gronenborn 2004). Following uncoating of the virion, viral dsDNA synthesis using host DNA polymerase(s) is primed via encapsidated short DNA primers complementary to the major common region, and from these dsDNA intermediates, host RNA polymerase then transcribes mRNAs encoding the viral proteins. Viral DNA replication is initiated by the M-Rep protein. There is experimental evidence for faba bean necrotic yellows virus (FBNYV) and BBTV that M-Rep has DNA cleavage and nucleotidyl transfer activity in vitro and initiates the replication of all genomic DNAs (Hafner et al., 1997, Timchenko et al., 1999, Timchenko et al., 2000). Each viral genome component contains a conserved nonanucleotide sequence flanked by inverted repeat sequences that potentially form a stem-loop structure and are a part of the viral origin of replication (ori). This sequence arrangement, including the loop-forming sequence containing the nonanucleotide 5′-TATTATT*AC-3′ or 5′-TAGTATT*AC-3′, is almost perfectly conserved in babuviruses and nanoviruses, respectively (an exception is the unclassified nanovirus parsley severe stunt associated virus, which has the same nonanucleotide as for babuviruses). The loop is cleaved at the position marked * to allow the initiation of replication. The non-coding regions (NCRs) of all genomic DNAs of a given nanovirid share this highly conserved stem-loop region (CR-SL) that also encompasses short repeated sequences (iterons) that are presumed to be binding sites for the M-Rep protein (Herrera-Valencia et al., 2006), the only viral protein essential for nanovirid replication. All other replication proteins including DNA polymerases are provided by the host cell. Viral DNA replication is enhanced by the action of Clink, a nanovirid-encoded cell cycle modulator protein which promotes cell cycle progression and transcription of S-phase relevant genes (Aronson et al., 2000, Lageix et al., 2007).
The movement protein of BBTV has been identified as a suppressor of RNA silencing (Amin et al., 2011, Niu et al., 2009).
The simultaneous presence of all genome components in a single infected host cell appears not to be required because nanovirid gene products may be capable of complementation at a supra-cellular level, illustrating a unique feature of these multipartite viruses (Sicard et al., 2019). Additionally, the relative proportions of the different genome components (termed the setpoint genome formula) attain stable, but different, rates in different host plant species (Sicard et al., 2013). Evidence of frequent interspecies and intraspecies recombination and reassortment events is present in nanovirid genomes (Grigoras et al., 2014, Stainton et al., 2012).
In addition to the genomic DNAs, a number of additional DNAs encoding Rep proteins have been found associated with some, but not all nanovirid infections. These additional DNAs, classified in alphasatellite species in different genera of the subfamily Nanoalphasatellitinae, family Alphasatellitidae (Briddon et al., 2018), are genetically very diverse and phylogenetically distinct from the DNA-R of nanoviruses and babuviruses. Tentative evidence suggests that these DNAs may modulate disease symptom expression (Gronenborn 2004).
Biology
Host range
Viruses of the individual species have narrow host ranges. Nanoviruses naturally infect only eudicotyledons, predominantly a limited range of leguminous plants (Fabaceae) (Vetten 2009, Franz et al., 1997, Yang et al., 2016), with the single exception of milk vetch dwarf virus (MDV) which also infects non-leguminous eudicots (Lal et al., 2020) and even the monocotyledon Lilium (Choi et al., 2019). Babuviruses have been reported only from a few monocotyledonous species from the Zingiberales (Musaceae and Zingiberaceae) (Thomas 2019) and Arecales (Arecaceae) (Pinili et al., 2013).
Viruses are restricted to the phloem tissue, and typically cause symptoms of stunting, reddening or yellowing of leaves, and sometimes leaf rolling and premature death (Vetten 2009, Thomas 2019, Magee 1927).
Transmission
In nature, nanovirids are transmitted by one or a limited number of aphid species in a persistent, non-propagative manner (Vetten et al., 2016). BBTV has been shown to translocate from the anterior midgut to the principal salivary glands, with no evidence of virus replication within the insect vector (Watanabe et al., 2016, Watanabe and Bressan 2013, Watanabe et al., 2013, Di Mattia et al., 2020). Using infectious, cloned genomic DNAs, it has been shown for faba bean necrotic stunt virus (FBNSV) that DNA-N is mandatory for aphid transmission, acting as a virus-encoded helper factor (Grigoras et al., 2018). The failure to transmit these viruses by aphids from purified virion preparations using membrane feeding techniques may result from the absence of the helper factor in purified virion preparations. Also, the absence of DNA-U1 results in greatly reduced aphid transmission efficiency. Interestingly, studies with FBNSV demonstrated reproducible differences in the genome formula of the virus in the bodies of both vector and non-vector species of aphids compared with the infected plant they had fed upon. These genome formula changes in the aphid were apparent in the early stages of the transmission cycle. The underlying reasons for these differences remain obscure (Sicard et al., 2015).
In some cases, nanovirids can be transmitted through vegetative planting material and grafting, but they are not transmitted through true seed or by conventional mechanical transmission. Several nanoviruses have been transmitted by biolistic bombardment using purified virions or cloned DNA components (Grigoras et al., 2014, Grigoras et al., 2009, Franz et al., 1999, Timchenko et al., 2006), but this strategy has not yet been successful for any babuvirus.
Geographical distribution
To date, nanovirids have only been reported from the Old World, and individual viruses vary greatly in their distribution range (Vetten 2009, Thomas 2019).
Among the babuviruses, BBTV is found widely distributed in Asia, Africa and the Pacific (Thomas 2019). ABTV has only been recorded from the Philippines and Malaysia (Sarawak) (Thomas 2019), and CBDV only from India (Mandal et al., 2013).
The nanovirus FBNYV is also widely distributed, occurring in several countries of West Asia and North and East Africa (Abraham et al., 2012, Kraberger et al., 2018) as well as Europe (Azerbaijan and Spain) (Grigoras et al., 2014), while subterranean clover stunt virus (SCSV) is reported only from Australia (Chu and Helms 1988) and pea necrotic yellow dwarf virus (PNYDV) from Central Europe (Gaafar et al., 2018, Gaafar et al., 2016).
Antigenicity
Virions are strongly immunogenic. Viruses within each of the two genera cross-react serologically, but there are no reports of serological relationships between members of the two genera (Abraham et al., 2010, Grigoras et al., 2010, Mandal et al., 2004, Sharman et al., 2008).
Derivation of names
Babuvirus: from banana bunchy top virus.
Nanoviridae, Nanovirus: from the Greek nanos, meaning "dwarf", referring to the observations that these plant viruses have the smallest known virions and genome segment sizes, and induce dwarfing symptoms in their hosts.
Genus demarcation criteria
Criteria used for differentiating the genera in this family (i.e. babuviruses from nanoviruses) are shown in Table 3 Nanoviridae and rely predominantly on molecular properties.
Table 3 Nanoviridae. Features distinguishing viruses of the genera Nanovirus and Babuvirus
| Features | Genus Nanovirus | Genus Babuvirus |
| Major hosts | Eudicotyledons | Monocotyledons |
| Major aphid vectors | Aphis craccivora and a few other legume-colonizing aphid species | Pentalonia and Micromyzus spp. |
| No. of genomic DNAs | 8 | 6 |
| Presence of specific DNAs | DNA-U1, DNA-U2, DNA-U4 | DNA-U3 |
| Length of genomic DNAs | 917–1046 nt | 1013–1114 nt |
| Nucleotide sequence identity of combined DNA-R, DNA-N, DNA-S, DNA-C and DNA-M | <62% (between babu- and nanoviruses) | |
Relationships within the family
Babuviruses and nanoviruses separate into distinct clades in phylogenetic analysis of concatenated sequences of the total genomic components of nanovirids (Figure 3 Nanoviridae). Babuviruses share significant levels of aa sequence similarity with nanoviruses only in the M-Rep (54–58%) and NSP (41–50%) (Vetten et al., 2012).
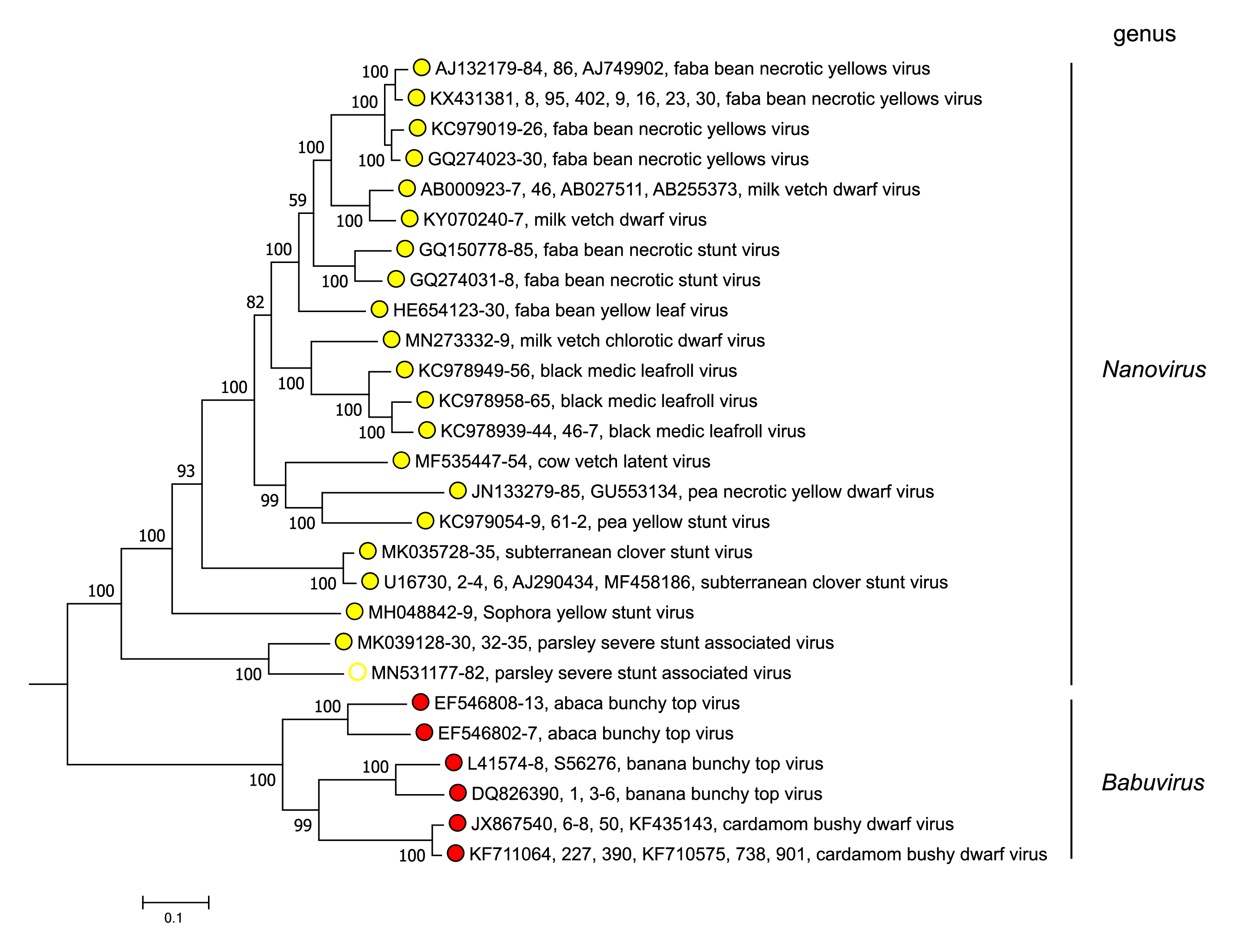 |
| Figure 3 Nanoviridae. Maximum likelihood dendrogram showing nucleotide sequence relationships in the concatenated DNA components (DNA-C, DNA-M, DNA-N, DNA-R and DNA-S, and DNA -U1, DNA-U2, DNA-U3 and DNA-U4 where present) from representative members of the genera Nanovirus and Babuvirus (family Nanoviridae). Vertical branch lengths are arbitrary and horizontal distances are proportional to the number of base substitutions per site (see scale bar). Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018), using the MUSCLE algorithm (Edgar 2004). The dendrograms were bootstrapped 500 times (scores are shown at nodes). This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
A comparison of concatemers of the total genomic components of nanovirids (incl. tentative nanoviruses) reveals 61 to 78% nt identity between members of different nanovirus species, 70 to 72% between members of different babuvirus species and 59% to 62% between members of the two genera, with a recently identified virus from parsley having borderline identities (60-64%) with members of assigned species of both genera and a similarly ambiguous phylogenetic position (Table 4 Nanoviridae (Resource page)).
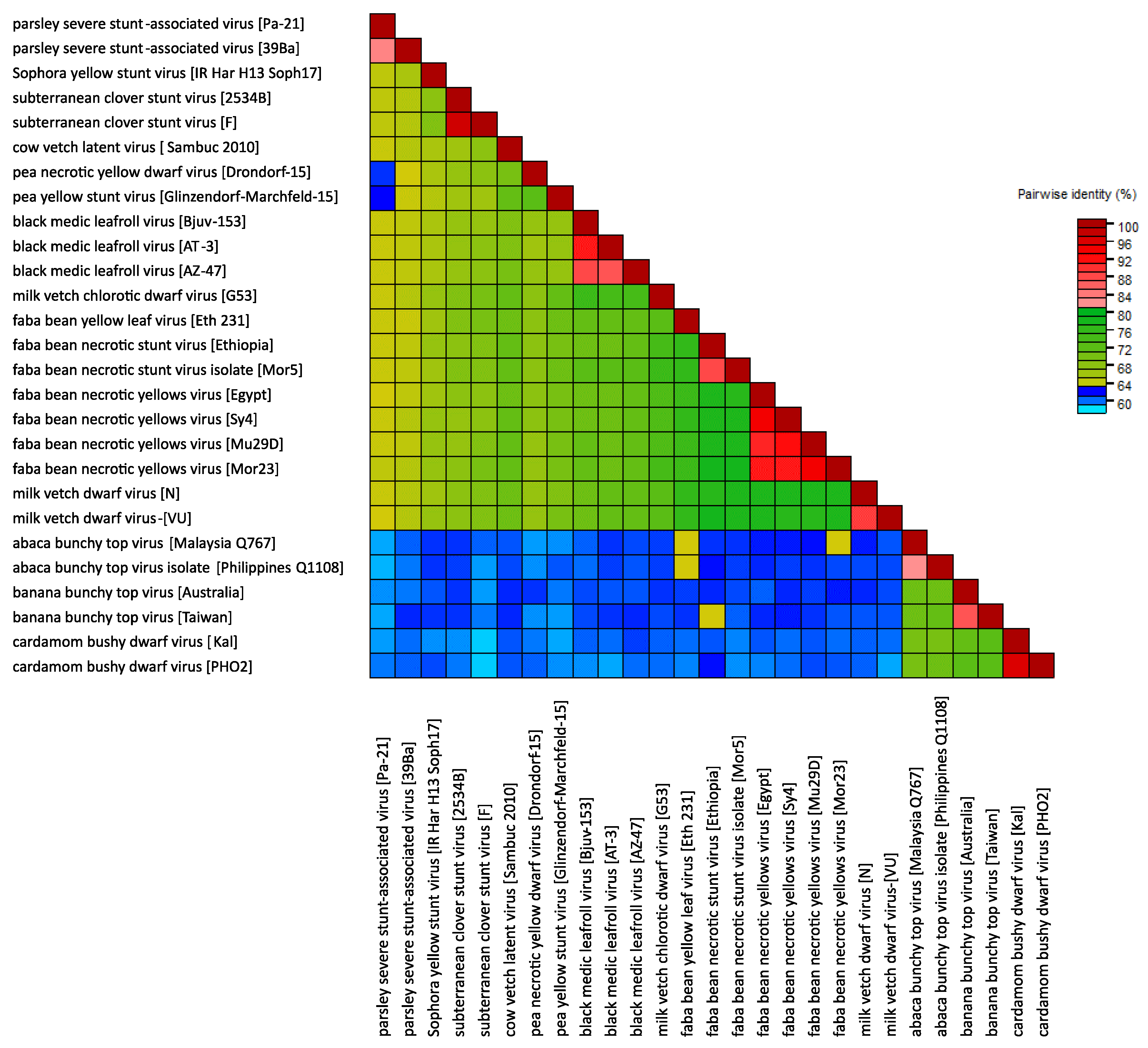 |
| Figure 4 Nanoviridae. Heat map illustrating pairwise distances between viruses in the family Nanoviridae, based on nucleotide sequences of their complete genomes (DNA-C, DNA-M, DNA-N, DNA-R and DNA-S, and DNA-U1, DNA-U2, DNA-U3 and DNA-U4 where present) and using the MUSCLE algorithm (Edgar 2004) in the program SDT 1.2 (Muhire et al., 2014). |
Relationships with other taxa
Nanovirids share properties with members of several other families of ssDNA viruses, and have recently been placed within the Cressdnaviricota, a virus phylum comprising families of Rep-encoding viruses with single-stranded, circular DNA genomes (Krupovic et al., 2020). All Rep proteins of members of nanovirid species have most of the amino acid motifs characteristic of Rep proteins of other ssDNA viruses including the plant-infecting geminiviruses (Gronenborn 2004). However, the nanovirid Rep proteins differ from those of geminiviruses (Zerbini et al., 2017) in being smaller (about 33 kDa vs. 40 kDa), having a slightly different dNTP-binding motif (GPQ/NGGEGKT), lacking the retinoblastoma-like protein (Rb)-binding motif (LxCxE) and sharing only 17 to 22% amino acid sequence identity with them. Moreover, geminiviruses differ in particle morphology (they are geminate), genome size, number and size of DNA components, mode of transcription, and in vector species. In comparison to nanovirids, members of the family Circoviridae possess closely related Rep proteins and morphologically similar virions, but infect vertebrates and crustaceans and have a much smaller genome (1.8–2.3 kb) that is monopartite, bidirectionally transcribed and contains only two genes (Breitbart et al., 2017). All these ssDNA viruses have a conserved nonanucleotide motif on the stem-loop sequence, with a conserved cleavage site, which is consistent with the operation of a rolling-circle model for DNA replication. Further information on eukaryotic circular Rep-encoding single-stranded (CRESS) DNA viruses and their relationship with nanovirids is provided by recent reviews by (Zhao et al., 2019) and (Kazlauskas et al., 2019).
The virus coconut foliar decay virus was formerly considered to belong to an unassigned species (Coconut foliar decay virus) in the family Nanoviridae, but has now been moved to the species Cofodevirus kokonas in the family Metaxyviridae.

