Family: Paramyxoviridae
Bert Rima, Anne Balkema-Buschmann, William G. Dundon, Paul Duprex, Andrew Easton, Ron Fouchier, Gael Kurath, Robert Lamb, Benhur Lee, Paul Rota and Linfa Wang
Corresponding author: Bert Rima ([email protected])
Edited by: Jens H. Kuhn and Stuart G. Siddell
Posted: September 2019, updated August 2020
PDF: ICTV_Paramyxoviridae.pdf
Summary
The family Paramyxoviridae consists of large enveloped RNA viruses infecting mammals and birds, or in some cases reptiles and fish (Table 1. Paramyxoviridae). Many paramyxoviruses are host-specific and several such as measles virus, mumps virus, Nipah virus, Hendra virus and several parainfluenza viruses are pathogenic for humans. Virus transmission is horizontal, mainly through direct contact and airborne routes; no vectors are known.
Table 1. Paramyxoviridae. Characteristics of members of the family Paramyxoviridae
| Characteristic | Description |
| Typical member | measles virus, Ichinose-B95a (AB016162), species Morbillivirus hominis |
| Virion | Enveloped, pleomorphic (mostly spherical) virions with a diameter of 300–500 nm enclosing a ribonucleoprotein |
| Genome | Negative-sense, non-segmented RNA genomes of 14.6–20.1 kb |
| Replication | Cytoplasmic, by the virus ribonucleoprotein complex, involves replication of antigenome and transcription of 6–8 positive-sense mRNAs |
| Translation | Cytoplasmic, by cellular machinery from capped and poly-adenylated mRNAs |
| Host range | Mammals, birds, fish and reptiles |
| Taxonomy | Realm Riboviria, kingdom Orthrnavirae, phylum Negarnaviricota, class Monjiviricetes, order Mononegavirales. Currently 9 subfamilies, 23 genera and 153 species |
Virion
Morphology
Virions are 150 nm or more (up to 500nm) in diameter, pleomorphic, but usually spherical in shape in vitreous ice. Virions consist of a lipid envelope surrounding a nucleocapsid. The envelope is derived directly from the host cell plasma membrane by budding and contains two transmembrane glycoproteins (Figure 1. Paramyxoviridae). These are present as homo-oligomers and form spike-like projections, 8–12 nm in length, spaced 7–10 nm apart (depending on virus genus affiliation). Also, depending on the genus, one or two additional transmembrane proteins may be present. One non-glycosylated membrane or matrix protein is associated with the inner face of the envelope. The virus nucleocapsid consists of negative-sense virus genome RNA and the nucleocapsid protein (N). The nucleocapsid has helical symmetry and is approximately 18 nm in diameter with a 7 nm pitch; its length can be up to 1,000 nm in viruses of some genera. The ribonucleoprotein (RNP) complex in the virion consists of the nucleocapsid together with the polymerase-associated or phosphoprotein (P) and the L protein (L, including RNA-directed RNA polymerase, capping and cap methylation activities) (Lamb and Parks 2007). Multiploid virions are found, although the vast majority of virions contain a single functional genome.
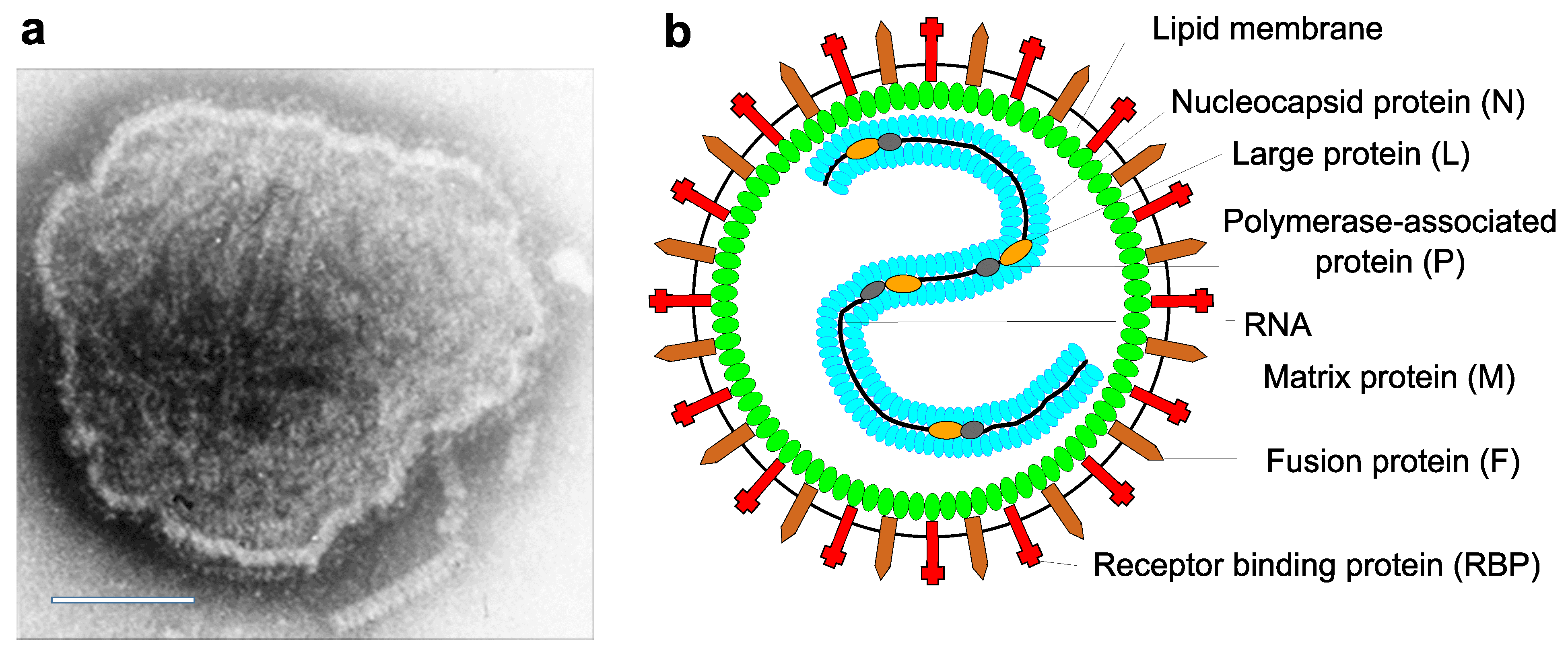 |
| Figure 1. Paramyxoviridae. Paramyxovirus virion structure. (A) Negative-contrast electron micrograph of intact measles virus particle (genus Morbillivirus). Scale bar = 100 nm. (B) Schematic diagram of paramyxovirus particle in cross-section. |
Physicochemical and physical properties
Virion Mr is about 500 ×106, and much greater for multiploid virions. Virion buoyant density in sucrose is 1.18–1.20 g cm−3. Virion S20,w is at least 1000S. Virions are very sensitive to heat, lipid solvents, ionic and non-ionic detergents, formaldehyde and oxidizing agents.
Nucleic acid
Virions contain a single molecule of linear, negative-sense, single stranded RNA that is not infectious alone but is infectious if the RNP complex is introduced into the cytoplasm. The RNA genome varies from 14,296 nucleotides for Antarctic penguin virus B to 20,148 nt for Pohorje Myodes paramyxovirus 1. Genomes of all viruses in the family Paramyxoviridae are multiples of 6 nt, which is a requirement for efficient replication (Calain and Roux 1993). Some virions may contain positive-sense RNA and so partial self-annealing of extracted RNA may occur. Intracellularly, or in virions, genome-length RNA is found exclusively encapsidated in ribonucleocapsids (RNPs). The genome RNA does not contain a 5′-cap, nor a covalently linked protein. The genome 3′-end is not polyadenylated.
Proteins
Members of the family Paramyxoviridae encode 6–10 proteins (5–250 kDa) of which several can be derived either from gene editing events in the P locus and an overlapping ORF in the P gene itself (Figure 2. .Paramyxoviridae). Virion proteins common to all genera include: three nucleocapsid-associated proteins, i.e., an RNA-binding nucleocapsid protein (N), a polymerase-associated phosphoprotein (P) and a large protein (L, including an RNA-directed RNA polymerase (RdRP), mRNA guanylyl- and methyltransferases, and methylation functions required for the capping of mRNAs), and three membrane-associated proteins, i.e., an unglycosylated inner membrane or matrix protein (M) and two glycosylated envelope proteins, comprising a fusion protein (F) and an attachment or receptor-binding protein (RBP, designated variably as HN, haemagglutinin-neuraminidase protein, H, haemagglutinin or G, glycoprotein). The F protein is synthesized within infected cells as a precursor (F0) that is activated following cleavage by cellular protease(s) to produce the virion disulfide-linked F1 and F2 subunits (order: N-F2-S-S-F1-C). Some viruses also encode putative non-structural proteins (C), a cysteine-rich protein that binds Zn2+ (V) that can be structural or non-structural depending on the virus, a small integral membrane protein (SH) and transmembrane proteins (tM). Some virus genomes, such as that of the fer-de-lance virus, contain transcription units encoding proteins with unidentified functions. Virion enzyme activities include the RNA-directed RNA polymerase and mRNA guanylyl- and methyltransferases functionally encoded in the L protein. Variously represented among the genera are neuraminidases associated with the RBP.
Lipids
Lipids in the virus envelope are derived from host cell plasma membrane.
Carbohydrates
Virions are composed of approximately 6% carbohydrate by weight; composition is dependent on the host cell. Fusion and RBP proteins are glycosylated by N-linked carbohydrate side chains.
Genome organization and replication
The genome organization is illustrated in Figure 2. .Paramyxoviridae for viruses representing 14 of the genera in the family.
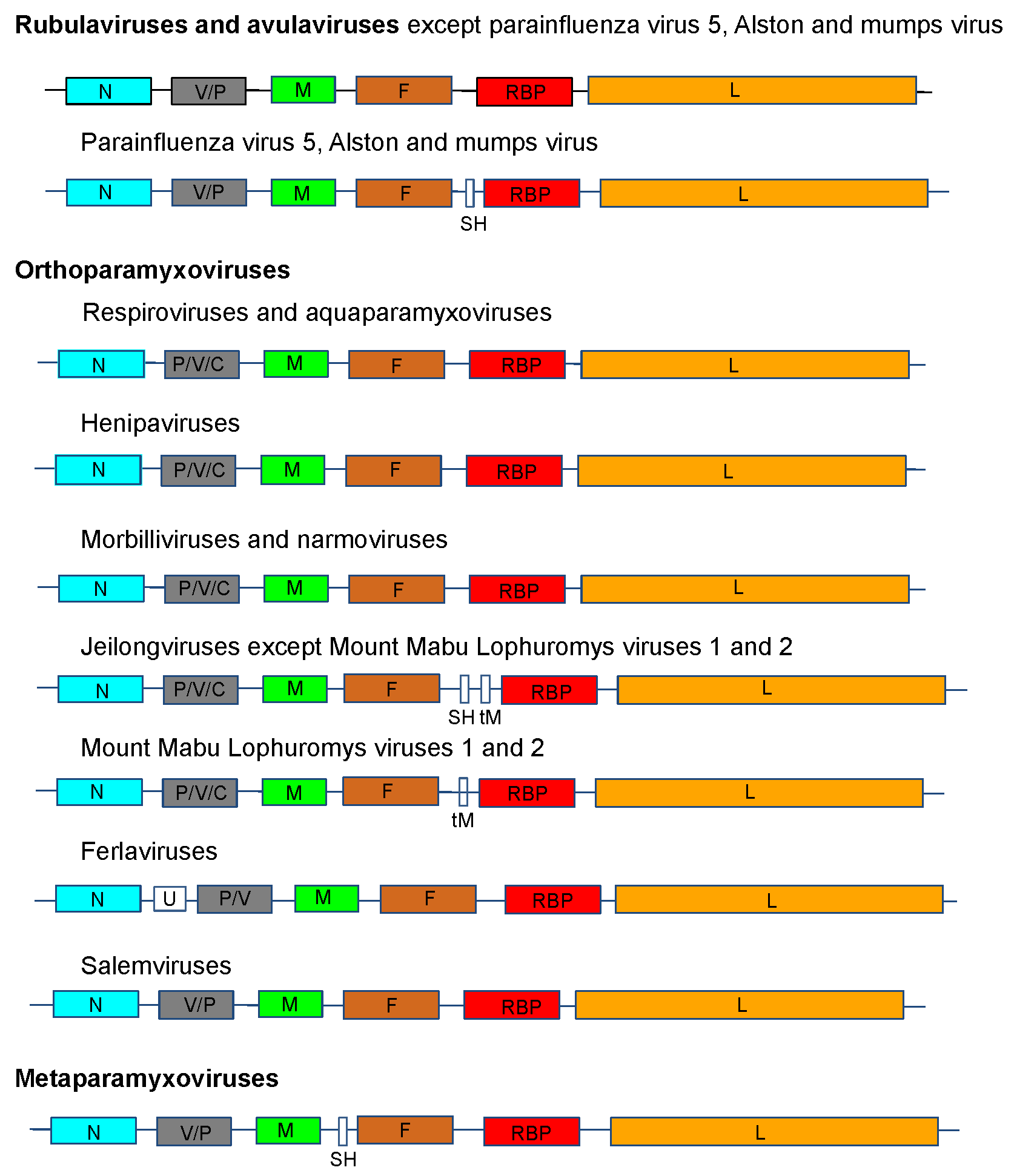 |
| Figure 2. .Paramyxoviridae. Genome organization (3′-to-5′) of viruses in the family Paramyxoviridae. Each box represents a separately encoded coding sequence; slashes indicate where multiple distinct ORFs are present within mRNA transcripts. Co-transcriptional editing leads to expression of the V or the P protein: the first shown is derived from the unedited sequence. The lengths of the boxes are approximately to scale although the non-coding sequences (NCS) are not to scale. Certain viruses express additional C proteins by the using multiple secondary translational start sites within the P gene. In human parainfluenza virus 1 and human parainfluenza virus 3 of the genus Respirovirus, the V ORF may be a non-expressed relic, the function of which may be partially compensated by an edited D protein. U is an additional transcription unit between the N and P genes in ferlavirus genomes. |
After attachment to cell receptors, virion entry is achieved by fusion of the virion envelope with the cell surface membrane. This can occur at neutral pH. Virus replication occurs in the cell cytoplasm and is thought to be independent of host nuclear functions. The genome is transcribed processively from the 3′-end by the virion-associated RdRP into 6–8 separate positive-sense mRNAs. Transcription is guided by short (10–13 nt) conserved gene start (GS) and gene end (GE) signals flanking the intergenic sequence. The mRNAs are capped by the guanylyl- and methyltransferase activities of the L protein and possess 3′-poly(A) tracts synthesized by reiterative copying of U tracts in GE sequence. Intergenic regions are highly conserved in length (3 nt) and sequence (CUU with few exceptions see Table 2. Paramyxoviridae for details) in the orthoparamyxoviruses and metaparamyxoviruses. Neither, the length or sequence of the intergenic sequences is conserved in avulavirus or rubulavirus genomes. RNA replication occurs through an intermediate, the antigenome, which is an exact positive-sense copy of the genome.
RNP assembly occurs in the cytoplasm and is tightly linked to RNA synthesis. RNPs are enveloped by budding at the cell surface plasma membrane at sites containing virion envelope proteins. Orthoparamyxovirus genomes contain 6–8 transcriptional elements that encode 7–11 proteins. Each element encodes a single mRNA with the exception of the P/V element. This element is transcribed into an exact copy mRNA (P or V mRNA, depending on genus) and into alternative versions in which the RNA transcriptase ‘stutters’ on the template at an editing motif midway down the element. This stuttering results in the insertion of one or more pseudo-templated G nucleotides (“RNA editing”) and shifts the reading frame to access alternative ORFs. The exact copy and edited mRNAs synthesize two alternative proteins, P and V, which have identical amino-terminal domains but due to the insertions of G residues have different carboxy-terminal domains. Other truncated, or chimeric, proteins (called I, W, or D, depending on the virus) can be produced by shifting into the third reading frame. The C ORF present in henipavirus, morbillivirus, narmovirus, jeilongvirus, aquaparamyxovirus and respirovirus genomes overlaps the P ORF and can initiate synthesis at an AUG codon that is accessed by ribosomal choice or at alternative start codons in the same ORF.
Biology
Paramyxoviruses have been conclusively identified only in vertebrates and mostly in mammals and birds, although they have recently also been detected in reptiles and fish, including boneless fish. Most viruses have a narrow host range in nature but can infect a broader range of cultured cells. Infection of cultured cells is generally lytic, but temperate or persistent infections are common in this family in vitro and in vivo. Other features of infection include the formation of inclusion bodies and syncytia. Host cell surface molecules reported to serve as receptors for the attachment for members of the family vary (Thibault et al., 2017). Respiroviruses, some rubulaviruses and all avulaviruses use sialoglycoproteins and glycolipids as receptors. The cell surface proteins signaling lymphocytic activation molecule family member 1 (SLAMF1, aka CD150) and nectin cell adhesion molecule 4 (nectin 4) are major receptors for measles virus and other morbilliviruses. Henipaviruses use ephrin B2 (EFNB2) and B3 (EFNB3) proteins as cellular entry receptors (Table 2. Paramyxoviridae).
Table 2. Paramyxoviridae. Receptor and receptor binding protein properties of paramyxoviruses
Orthoparamyxovirinae
| Genus | Virus | RBP name / amino acid residues# | Sequence at start of RBP propeller blade 2* | Cell receptor | Intergenic trinucleotides
|
| Aquaparamyxovirus | Atlantic salmon paramyxovirus | HN 576 | NRKSCS | ? probably neuraminic acid | CUU + CAU (F-HN) |
Aquaparamyxovirus
| Pacific salmon paramyxovirus | HN 578 | NRKSCS | ? probably neuraminic acid | CUU |
| Ferlavirus | fer-de-lance virus | HN 564 | NRKSCS | ? probably neuraminic acid | CCU(3x)+ CUU(4x) alternating |
| Jeilongvirus | Beilong virus | “G” 734 | NRRSCT | ? | CUU |
| Jeilongvirus | Tailam virus | “G” 1052 | NRRSCT | ? | CUU |
| Jeilongvirus | J-virus | “G” 709 | NRRSCS | ? | CUU |
| Jeilongvirus | Pohorje Myodes paramyxovirus 1 | “G” 1589 | NRRSCT | ? | CUU |
| Jeilongvirus | Mount Mabu Lophuromys virus 1 | “G” 854 | NRKSCT | ? | CUU |
| Jeilongvirus | Mount Mabu Lophuromys virus 2 | “G” 810 | NRKSCS | ? probably neuraminic acid | CUU |
| Jeilongvirus | Shaan virus | HN 588 | NRKSCS | ? probably neuraminic acid | CUU + CGU (F-SH) |
| Henipavirus | Hendra virus | G 604 | TIHHCS | EFNB2/3 | CUU |
| Henipavirus | Nipah virus | G 420 | TVYHCS | EFNB2/3 | CUU |
| Henipavirus | Cedar virus | G 622 | QVINCV | EFNB2 | CUU |
| Henipavirus | Mòjiāng virus | G 625 | IINSCA | protein? | CUU |
| Henipavirus | Ghana virus | G 632 | NYHSCT | EFNB2 | CUU + CUG (F-G) |
| Morbillivirus | measles virus wt | H 617 | DFSNCM | SLAMF1/NECTIN4 | CUU + CGU (H-L) |
| Morbillivirus | measles Ed-Zag vac | H 617 | DLSNCM | SLAMF1/NECTIN4/CD46 | CUU + CGU (H-L) |
| Morbillivirus | canine distemper virus | H 607 | KTKVCT | SLAMF1/NECTIN4 | CUU + CUA (H-L) |
| Morbillivirus | canine distemper virus vaccine | H 607 | KAKVCT | SLAMF1/NECTIN4/? | CUU + CUA (H-L) |
| Morbillivirus | phocine distemper virus | H 607 | NTKICT | SLAMF1/NECTIN4t | CUU + CUA (H-L) |
| Morbillivirus | rinderpest virus | H 609 | ELETCM | SLAMF1/NECTIN4 | CUU + CGU (H-L) |
| Morbillivirus | peste des petits ruminants virus | H 609 | DYRSCL | SLAMF1/NECTIN4 | CUU |
| Morbillivirus | dolphin morbillivirus | H 604 | GLNFCL | SLAMF1/ NECTIN4 | CUU |
| Morbillivirus | feline morbillivirus | H 595 | GMESCT | SLAMF1/ NECTIN4 | CUU + CUA (M-F) |
| Narmovirus | Nariva virus | “H” 657 | AYDGCA | protein? | CUU |
| Narmovirus | Mossman virus | “G” 632 | VFDGCS | protein? | CUU + CGU (F-H) |
| Narmovirus | bank vole virus 1 | “G” 625 | LRDSCT | protein? | CUU + CUA (P-M and F-H and L-t); CAU (M-F) |
| Narmovirus | Tupaia paramyxovirus | “H” 665 | NLRDCS | protein? | CUU |
| Respirovirus | human parainfluenza virus 1 | HN 575 | NRKSCS | Neuraminic acid | CUU + CGU (P-M) |
| Respirovirus | Sendai virus | HN 575 | NRKSCS | Neuraminic acid | CUU + CCC (HN-L) |
| Respirovirus | giant squirrel virus | HN 574 | NRKSCS | ? probably neuraminic acid | CUU +CAU (HN-L) |
| Respirovirus | human parainfluenza virus 3 | HN 572 | NRKSCS | Neuraminic acid | CUU |
| Respirovirus | bovine parainfluenza virus 3 | HN 572 | NRKSCS | Neuraminic acid | CUU |
| Respirovirus | porcine parainfluenza virus 1 | HN 576 | NRKSCS | ? probably neuraminic acid | CUU |
| Respirovirus | caprine parainfluenza virus 3 | HN 574 | NRKSCS | ? probably neuraminic acid | CUU |
| Salemvirus | Salem virus | “G” 620 | LSGKCT | protein? | CUU + CCU(P-M) + CGU (F-G) |
Metaparaymyxovirinae
| Genus | Virus | RBP name / amino acid residues# | Sequence at start of RBP propeller blade 2* | Cell receptor | Intergenic trinucleotides
|
| Synodonvirus | Wēnlǐng triplecross lizardfish paramyxovirus | “HN”621 | PAPSCP | protein? | CUU + CAUCUU (F-HN) |
Rubulavirinae
| Genus | Virus | RBP name/ amino acid residues# | Sequence at start of RBP propeller blade 2* | Cell receptor |
| Orthorubulavirus | mumps virus | HN 582 | NRKSCS | Neuraminic acid |
| Orthorubulavirus | La Piedad Michoacán Mexico virus | HN 576 | NRKSCS | ? probably neuraminic.acid |
| Orthorubulavirus | Mapuera virus | HN 582 | NRKSCS | ? probably neuraminic acid |
| Orthorubulavirus | simian virus 41 | HN 568 | NRKSCS | ? probably neuraminic acid |
| Orthorubulavirus | human parainfluenza virus 2 | HN 571 | NRKSCS | ? probably neuraminic acid |
| Orthorubulavirus | human parainfluenza virus 4 | HN 579 | NRKSCS | ? probably neuraminic acid |
| Orthorubulavirus | parainfluenza virus 5 | HN 532 | NRKSCS | Neuraminic acid |
| Orthorubulavirus | Alston virus | HN 565 | NRKSCS | ? probably neuraminic acid |
| Pararubulavirus | Menangle virus | “HN” 595 | PVRTCS | protein? |
| Pararubulavirus | Tioman virus | “HN” 593 | QARGCS | protein? |
| Pararubulavirus | Teviot virus | “HN” 595 | QTRGCS | protein? |
| Pararubulavirus | Achimota virus 1 | “HN” 595 | VTYQCS | protein? |
| Pararubulavirus | Achimota virus 2 | “HN” 583 | FRRGCS | protein? |
| Pararubulavirus | Hervey virus | “HN” 543 | PKRSCS | protein? |
| Pararubulavirus | Tuhoko virus 1 | “HN” 580 | WLRSCS | protein? |
| Pararubulavirus | Tuhoko virus 2 | “HN” 588 | VSRQCS | protein? |
| Pararubulavirus | Tuhoko virus 3 | “HN” 582 | RLYHCS | protein? |
| Pararubulavirus | Sosuga virus | “HN” 582 | RLYHCS | protein? |
Avulavirinae
| Genus | Virus | RBP name/ amino acid residues | Sequence at start of RBP propeller blade 2* | Cell receptor |
| Metaavulavirus | avian paramyxovirus 2 | HN 580 | NRKSCS | ? probably neuraminic acid |
| Metaavulavirus | avian paramyxovirus 5 | HN 574 | NRKSCS | ? probably neuraminic acid |
| Metaavulavirus | avian paramyxovirus 6 | HN 613 | NRKSCS | ? probably neuraminic acid |
| Metaavulavirus | avian paramyxovirus 7 | HN 569 | NRKSCS | ? probably neuraminic acid |
| Metaavulavirus | avian paramyxovirus 8 | HN 577 | NRKSCS | ? probably neuraminic acid |
| Metaavulavirus | avian paramyxovirus 10 | HN 575 | NRKSCS | ? probably neuraminic acid |
| Metaavulavirus | avian paramyxovirus 11 | HN 583 | NRKSCS | ? probably neuraminic acid |
| Metaavulavirus | avian paramyxovirus 14 | HN 580 | NRKSCS | ? probably neuraminic acid |
| Metaavulavirus | avian paramyxovirus 15 | HN 579 | NRKSCS | ? probably neuraminic acid |
| Metaavulavirus | avian paramyxovirus 20 | HN 574 | NRKSCS | ? probably neuraminic acid |
| Orthoavulavirus | avian paramyxovirus 1 (NDV) | HN 571 | NRKSCS | Neuraminic acid |
| Orthoavulavirus | avian paramyxovirus 9 | HN 579 | NRKSCS | ? probably neuraminic acid |
| Orthoavulavirus | avian paramyxovirus 12 | HN 614 | NRKSCS | ? probably neuraminic acid |
| Orthoavulavirus | avian paramyxovirus 13 | HN 579 | NRKSCS | ? probably neuraminic acid |
| Orthoavulavirus | avian paramyxovirus 16 | HN 618 | NRKSCS | ? probably neuraminic acid |
| Orthoavulavirus | Antarctic penguin virus A | HN 599 | NRKSCS | ? probably neuraminic acid |
| Orthoavulavirus | Antarctic penguin virus B | HN 591 | NRKSCS | ? probably neuraminic acid |
| Orthoavulavirus | Antarctic penguin virus C | HN 587 | NRKSCS | ? probably neuraminic acid |
| Orthoavulavirus | avian paramyxovirus 21 | HN 567 | NRKSCS | ? probably neuraminic acid |
| Paraavulavirus | avian paramyxovirus 3 | HN 577 | NRKSCS | ? probably neuraminic acid |
| Paraavulavirus | avian paramyxovirus 4 | HN 569 | NRKSCS | ? probably neuraminic acid |
# The nomenclature for RBP (G, H or HN) used in the accessions in the data bank submissions is shown in quotation marks.
* The canonical NRKSCS sequence at the start of propeller blade 2 (Langedijk et al., 1997) is shown in bold lettering; in the Avulavirinae and Rubulavirinae intergenic sequences vary widely in length and sequence and hence are not recorded in the Table.
Nucleocapsids associate with virus membrane proteins at the plasma membrane and are enveloped by budding out at the membrane.
Transmission of paramyxoviruses is horizontal, mainly through airborne and direct contact routes; no vectors are known. Paramyxovirus infection typically begins in the respiratory tract and may remain at that site (e.g., human parainfluenza virus 1 [HPIV-1]) or spread to secondary sites (e.g., lymphoid and endothelial tissues for measles virus (MV) (Griffin 2007), the parotid gland, CNS and endothelial tissues for mumps virus (MuV) (Carbone and Rubin 2007) or lung and CNS for Hendra virus (HeV) and Nipah virus (NiV) (Eaton et al., 2007). In general, paramyxovirus infections are limited, and eliminated, by host immunity. However, virus can sometimes be shed for periods of weeks or months in healthy and, especially, in immunocompromised individuals. Latent infection is unknown. However, long-term persistent infection is known for several morbilliviruses such as MV in subacute sclerosing panencephalitis, a rare complication that involves persistence of a defective measles virus in the CNS for periods of, on average, 8 years. Old dog distemper can involve persistence of defective or fully infectious canine distemper virus for weeks or months in healthy and, especially, immunocompromised animals. Feline morbillivirus has been shown to be shed for long periods from the kidneys of cats. The recurrence of neurological manifestations has also been noted in NiV patients more than 4 years after recovery from acute encephalitis (Eaton et al., 2007).
Antigenicity
The RBP and F proteins are of primary importance in inducing virus-neutralizing antibodies and immunity against reinfection. Antibodies to N and, variably, to other virus proteins also are induced by infection. Following processing into small peptides the virus proteins also stimulate cell-mediated immune responses.
Derivation of names
Avula: from avian and rubula
Cynoglossusvirus: from the genus Cynoglossus of the fish from which the virus sequence was obtained
Henipa: from Hendra and Nipah viruses, the first isolates assigned to this genus
Hoplichthysvirus: from the genus Hoplichthys of the fish from which the virus sequence was obtained
Meta: from Greek meta, meaning “after, beyond”.
Morbilli: from Latin morbillus, diminutive of morbus, “disease”.
Ortho: from Greek orthos, “straight”.
Paramyxo: from Greek para, “by the side of”, and myxa, “mucus”.
Pneumo: from Greek pneuma, “breath”.
Respiro: from Latin respirare, “respire, breathe”.
Rubula: from Rubula inflans – old name for the disease mumps from Latin Rubula, red; inflans, swelling or puffing up.
Scoliodonvirus: from binomial name Scoliodon macrorhynchos (Bleeker 1852) of Pacific spadenose shark from which the virus sequence was obtained
Subfamily, genus and species demarcation criteria
The current paramyxovirus taxonomic structure is based on a comparison of complete L protein amino acid sequences. The Paramyxoviridae Study Group decided to use this as a sole criterion on the basis of the likely monophyly of this large and complex virus protein (Wolf et al., 2018, Dolja and Koonin 2018) consequential upon the ICTV decision to classify viruses even if only known from their genome sequences (Simmonds et al., 2017) . The genetic-based classification reflected previous classifications based on biological characteristics, which are unlikely to be known for all the new paramyxovirus sequences that have become available (Rima et al., 2018). Four subfamilies have been established on the basis of their genetic distance from the node distinguishing the family Paramyxoviridae from the Sunviridae, which is the closest related outlier family. These distances (substitutions per site) are respectively 0.64 for the Metaparamyxovirinae, 0.80 for the Avulavirinae, 0.82 for the Orthoparamyxovirinae and 0.90 for the Rubulavirinae.
Relationships within the family
Phylogenetic analysis of complete L protein amino acid sequences (Figure 3. Paramyxoviridae) supports the classification of paramyxoviruses into four subfamilies and fourteen genera based on genetic distances; in addition, three viruses are members of species that are not assigned to a genus or subfamily
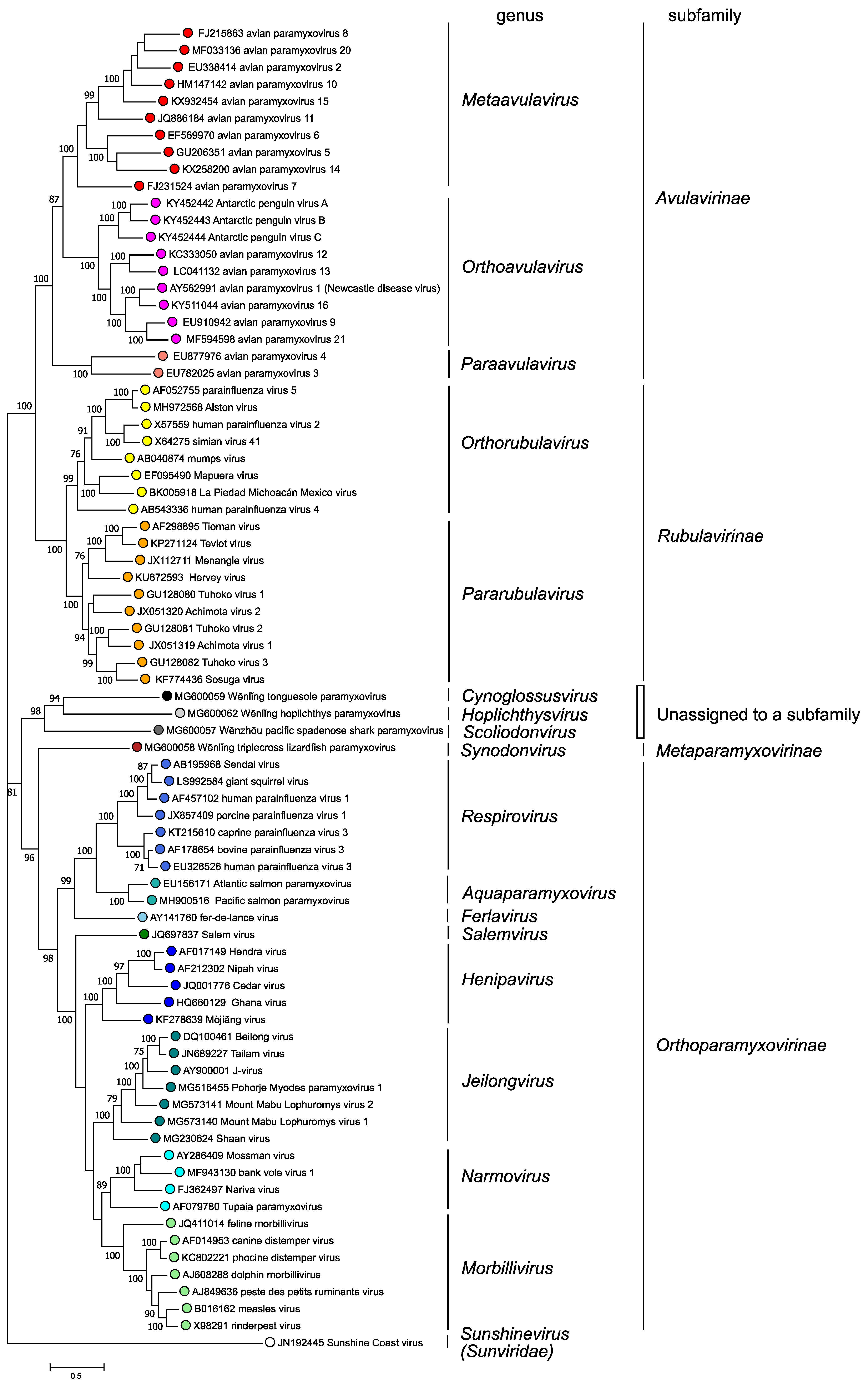 |
| Figure 3. .Paramyxoviridae. Phylogenetic analysis of complete L protein amino acid sequences of members of the family Paramyxoviridae. Complete L protein amino acid sequences were aligned by Clustal W with gap generation penalties of 5 and extension penalties of 1 in both multi and pairwise alignments. The evolutionary history was inferred by using the Maximum Likelihood method and JTT matrix-based model. The tree with the highest log likelihood (-258124.74) is shown. The percentage of 500 trees in which the associated taxa clustered together is shown next to the branches where this was > 70%. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 78 amino acid sequences. There were a total of 2745 positions in the final dataset. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018). This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
The member viruses of the family Paramyxoviridae have a similar strategy of gene expression and replication and gene order to those of other mononegaviruses, specifically filoviruses and rhabdoviruses.

