Family: Plectroviridae
Petar Knezevic and Evelien M. Adriaenssens
The citation for this ICTV Report chapter is the summary published as Knezevic et al., (2021):
ICTV Virus Taxonomy Profile: Plectroviridae, Journal of General Virology, 102 (5): 001597
Corresponding author: Petar Knezevic ([email protected])
Edited by: Evelien M. Adriaenssens and Stuart G. Siddell
Posted: April 2021
PDF: ICTV_Plectroviridae.pdf
Summary
Members of the family Plectroviridae produce particles that are non-enveloped rigid rods (70−280 × 10−16 nm) (Table 1. Plectroviridae). The supercoiled, circular, single stranded DNA genome of about 4.5−8.3 kb, encodes 4−13 proteins. Viruses of this family infect cell wall-less bacteria, adsorbing to the bacterial surface, replicating their DNA by a rolling circle mechanism or transposition and releasing progeny from cells by extrusion, without killing the host.
Table 1.Plectroviridae. Characteristics of members of the family Plectroviridae
| Characteristic | Description |
| Example | Acholeplasma phage MV-L1 (X58839), species Plectrovirus L51 |
| Virion | Non-enveloped rigid rods; 10−16 nm in diameter, 70−280 nm in length |
| Genome | 4.5−8.3 kb, supercoiled, circular, positive-sense single-stranded DNA; 4−13 encoded proteins |
| Replication | Rolling-circle or transposition |
| Translation | From mRNAs |
| Host range | Cell wall-less bacteria |
| Taxonomy | Realm Monodnaviria; kingdom: Loebvirae; phylum Hofneiviricota; class Faserviricetes, order Tubulavirales; the family Plectroviridae includes four genera and six species |
Virion
Morphology
The virions of members of the family Plectroviridae are rigid, asymmetric, non-enveloped, nearly straight rods with one end rounded, and the other more variable (Figure 1. Plectroviridae). Acholeplasma phages are 70–90 nm long and 14−16 nm in diameter, whereas several phages that infect Spiroplasma are 230–280 nm long and 10–16 nm wide (Bruce et al., 1972, Liss and Maniloff 1973, Liss and Cole 1982), being shorter and wider than members of the families Inoviridae and Paulinoviridae, the other families in the order Tubulavirales. Images of negatively-stained virions suggest that plectrovirus particles have 4±2 nm hollow cores. Optical diffraction images of Acholeplasma phage MV-L1 suggest morphological units arranged with two-fold rotational and 5.6-fold screw symmetry (Day 2012).
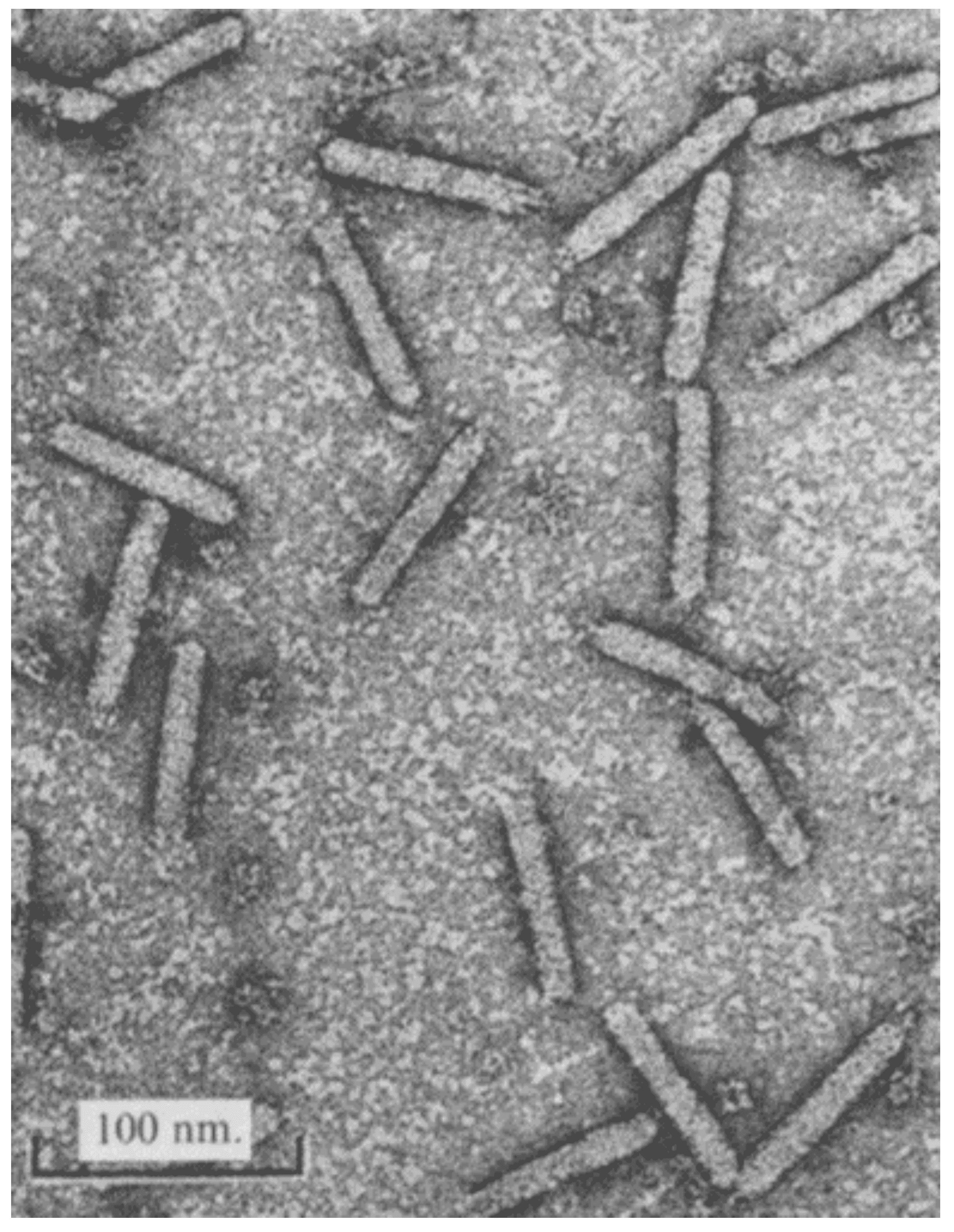 |
| Figure 1. Plectroviridae. Electron micrograph of purified preparation of Acholeplasma phage MV-L1, negatively-stained with 2% uranyl acetate (from (Bruce et al., 1972) with permission). |
Physicochemical and physical properties
The buoyant density of members of the genus Vespertiliovirus in metrizamide is 1.29 g/cm3 and in CsCl 1.39 g/cm3 (Melcher and Comer 2011). Acholeplasma phage MV-L1 (genus Plectrovirus) is resistant to detergents and stable during 30 minutes at 60 oC, but sensitive to chloroform. Similar properties have been confirmed for vespertilioviruses, which are also resistant to nonionic detergents such as Nonidet P-40 and Triton X-100, but are sensitive to chloroform and ether (Gourlay 1974).
Nucleic acid
The genome is a positive-sense (+) supercoiled circular ssDNA molecule (Figure 2. Plectroviridae) ranging from 4.5 kb for Acholeplasma phage MV-L1 to 8.3 kb for Spiroplasma phage 1-R8A2B and encoding between 4 and 13 proteins. The G+C content varies from 22.2% for Spiroplasma phage SkV1CR23x to 33.3% for Acholeplasma phage MV-L1.
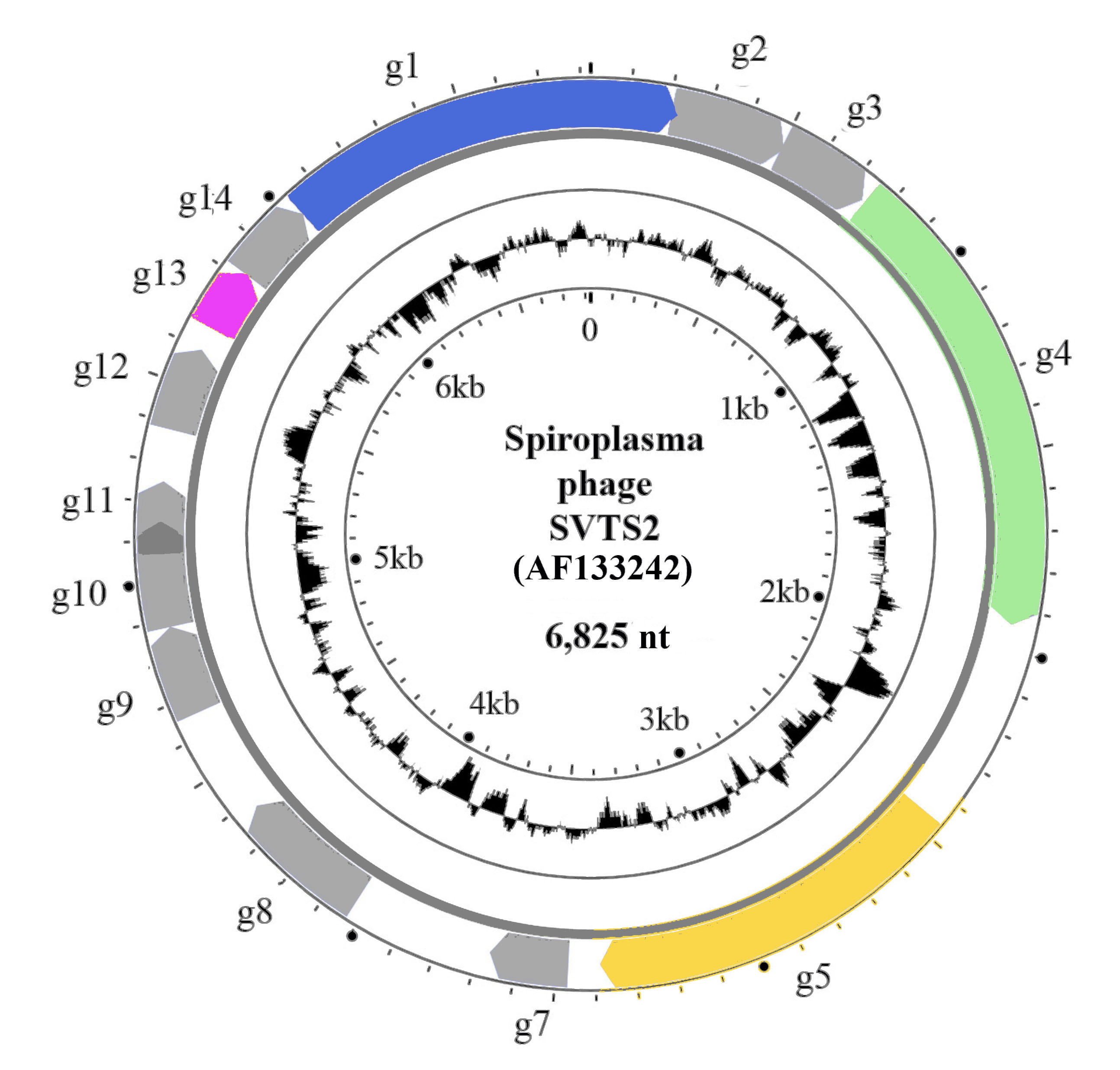 |
| Figure 2. Plectroviridae. Genome organisation of Spiroplasma phage SVTS2. The outer circle shows the circular genome (AF133242) with genes g1–g5 and g7–14 indicated by coloured arrows (g10 and g11 overlap; there is no g6). Genes with known products are coloured: g1- adhesion protein (CoaA); g4 - maturation (morphogenesis, assembly) protein; g5 - replication protein and g13 - major coat protein (CoaB). The inner circle represents GC content. |
Proteins
The mass of the major coat protein is 7.5 kDa for Spiroplasma phage 1-R8A2B (Renaudin and Bové 1994) but 19 kDa for Acholeplasma phage MV-L51, as assessed by gel mobility. The major coat protein has a strong tendency to aggregate.
Lipids
Not reported.
Carbohydrates
Not reported.
Genome organization and replication
Members of the family Plectroviridae encode from four to 13 proteins. Apart from the major coat protein, the DDE-type transposase in members of the genus Vespertiliovirus and the replication protein in members of the genus Suturavirus, the function of plectrovirus proteins is unknown. As a consequence, it is not known if members of the family Plectroviridae have a modular genome organization similar to that of members of the family Inoviridae. The receptors for these viruses are surface molecules or structures on the host cells. For Acholeplasma phage MV-L1, the cell-surface receptors are protein-lipoglycane molecules (Al-Shammari and Smith 1982). The DNA genome replicates by a rolling-circle mechanism in members of the genus Plectrovirus and, by inference, for members of the genus Suturavirus. Phages from the genus Vespertilliovirus possess a transposase gene instead of the p2 equivalent (replication initiation protein) of Escherichia phage M13 (Inoviridae) and replicate by a transposition mechanism with the encapsidated genome representing a circular transposition intermediate (Melcher et al., 1999). A specific trait of phages in the genera Suturavirus and Vespertiliovirus is that UGA in mRNAs is not a stop codon, but as in the host bacterium encodes tryptophan (Renaudin et al., 1990).
Virions assemble and are released at the membrane by extrusion, the host cells continuing to divide.
Biology
Hosts for phages belonging to the family Plectroviridae are cell wall-less bacteria from the genera Acholeplasmaa and Spiroplasma. As for other members of the order Tubulavirales, phages in the family Plectroviridae are neither obligatory lytic nor typical temperate and establish a chronic productive infection. Infected cells continuously release virions by extrusion while maintaining cellular integrity and viability. Acholeplasma phage MV-L1 and Spiroplasma phage SVTS2 can form plaques on appropriate host strains (Sha et al., 1995). Turbid plaques are produced in some superinfected mycoplasma strains with latent prophages. Members of the genus Vespertiliovirus, encoding a transposase, alter spiroplasma genomes by insertion within active genes, so destroying their function, providing targets for site-specific recombination, mediating deletions of sequences adjacent to their integration sites, and by providing targets for homologous recombination, leading to genome inversions (Melcher et al., 1999). Furthermore, for members of the genus Suturavirus, integration of viral DNA sequences into host chromosomal and extra-chromosomal DNA can cause resistance to reinfection with the same virus (Sha et al., 1995).
Genus demarcation criteria
Members of the same genus share considerable similarity of DNA sequences (BLASTn), and >50% similarity (identity × query coverage) of the major coat protein (CoaB) and maturation protein sequences (BLASTp).
Derivation of names
Plectroviridae, Plectrovirus: from the Latin plectrum, plectri meaning a small stick.
Suturavirus: from the Latin sutura, suturae meaning a stitch of sewing.
Vespertiliovirus: from the Latin vespertilio, vespertilionis meaning bat.
Relationships within the family
Members of the family Plectroviridae have similar genome organizations. Their phylogenetic relationships are presented in Figure 3. Plectroviridae.
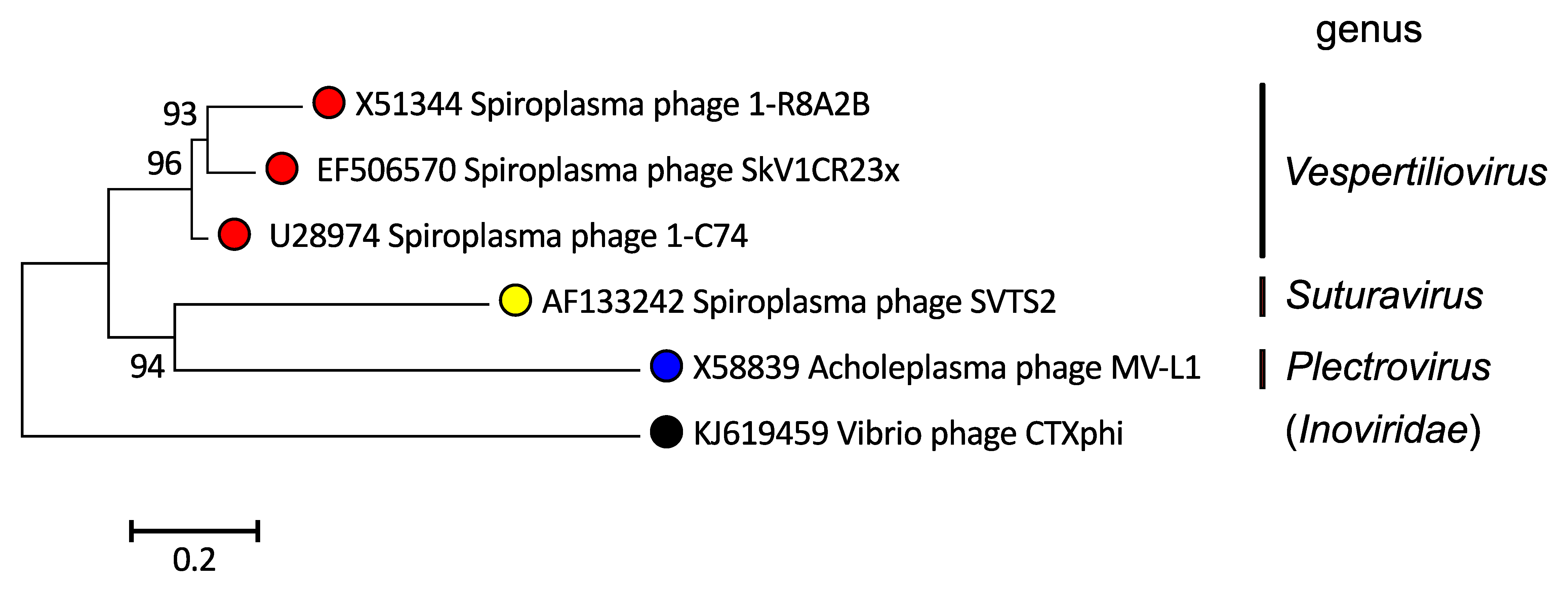 |
| Figure 3. Plectroviridae. A phylogenetic tree based on whole genome sequences of members of the family Plectroviridae was inferred using the Neighbor-Joining method (Saitou and Nei 1987). The tree was rooted using Vibrio phage CTXphi (Inoviridae) genome as an outlier. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (Felsenstein 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura 2-parameter method (Kimura 1980) and are in the units of the number of base substitutions per site. Codon positions included were 1st+2nd+3rd+Noncoding. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 11763 positions in the final dataset. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018, Stecher et al., 2020). This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Members of the family Plectroviridae are morphologically similar to members of the other families in the order Tubulavirales (Inoviridae and Paulinoviridae), and have the same nucleic acid type (positive-sense single-stranded DNA), replication cycle and other properties.

