Family: Portogloboviridae
David Prangishvili, Ying Liu and Mart Krupovic
The citation for this ICTV Report chapter is the summary published as Prangishvili et al., (2021):
ICTV Virus Taxonomy Profile: Portogloboviridae, Journal of General Virology, 102 (6): 001605
Corresponding authors: David Prangishvili ([email protected]) and Mart Krupovic ([email protected])
Edited by: Mart Krupovic and Stuart G. Siddell
Posted: April 2021
PDF: ICTV_Portogloboviridae.pdf
Summary
Portogloboviridae is a family of viruses with circular, double-stranded DNA genomes of about 20 kbp (Table 1. Portogloboviridae). Their icosahedral virions have a diameter of 87 nm, and consist of an outer protein shell, inner lipid layer and a nucleoprotein core wound up into a spherical coil. Portogloboviruses infect hyperthermophilic archaea of the genus Saccharolobus, order Sulfolobales and are presumably nonlytic. Portogloboviruses encode mini-CRISPR arrays which they use to compete against other co-infecting viruses.
Table 1. Portogloboviridae. Characteristics of members of the family Portogloboviridae
| Characteristic | Description |
| Example | Sulfolobus polyhedral virus 1 (KY780159), species Alphaportoglobovirus beppuense |
| Virion | Icosahedral virions, 87 nm in diameter, consisting of an outer protein shell, inner lipid layer and a nucleoprotein core wound up into a spherical coil |
| Genome | Circular, double-stranded DNA genomes of about 20 kbp |
| Replication | Viral replication occurs by chronic infection without observable cell lysis |
| Translation | Not characterized |
| Host range | Hyperthermophilic archaea of the genus Saccharolobus, order Sulfolobales |
| Taxonomy | One genus, two species |
Virion
Morphology
Virions are icosahedral, with a diameter of about 87 nm from vertex to vertex and 83 nm from facet to facet (Figure 1A.Portogloboviridae, 1B.Portogloboviridae), and consist of three structural units: (a) the outer icosahedral protein shell, (b) the subcapsomer proteins and lipid membrane and (c) a circular nucleoprotein wrapped into a spherical core (Figure 1C. Portogloboviridae) (Liu et al., 2017, Wang et al., 2019). The outer shell carries capsid proteins VP4 and VP10, which assemble into a T = 43 icosahedral shell. The five-fold vertices of the capsid are decorated with the vertex complexes extending above the capsid surface and potentially involved in receptor binding (Wang et al., 2019). The internal membrane of the virus also displays an icosahedral shape, presumably due to the presence of an intricate array of subcapsomer proteins that span the space between the membrane and the external icosahedral capsid (Wang et al., 2019, Baquero et al., 2020). The nucleoprotein filament, formed by multiple copies of the nucleocapsid protein VP1 and circular dsDNA, is condensed into a globular core with the characteristic appearance of concentric rings, seen in the axial view, and striations, seen in the side view (Figure 1B.Portogloboviridae) (Liu et al., 2017, Wang et al., 2019). The viral dsDNA is stored in the capsid in the A-form (11.0 bp/turn) (Wang et al., 2019), as previously observed for filamentous archaeal viruses of the realm Adnaviria (Baquero et al., 2020, Wang et al., 2020).
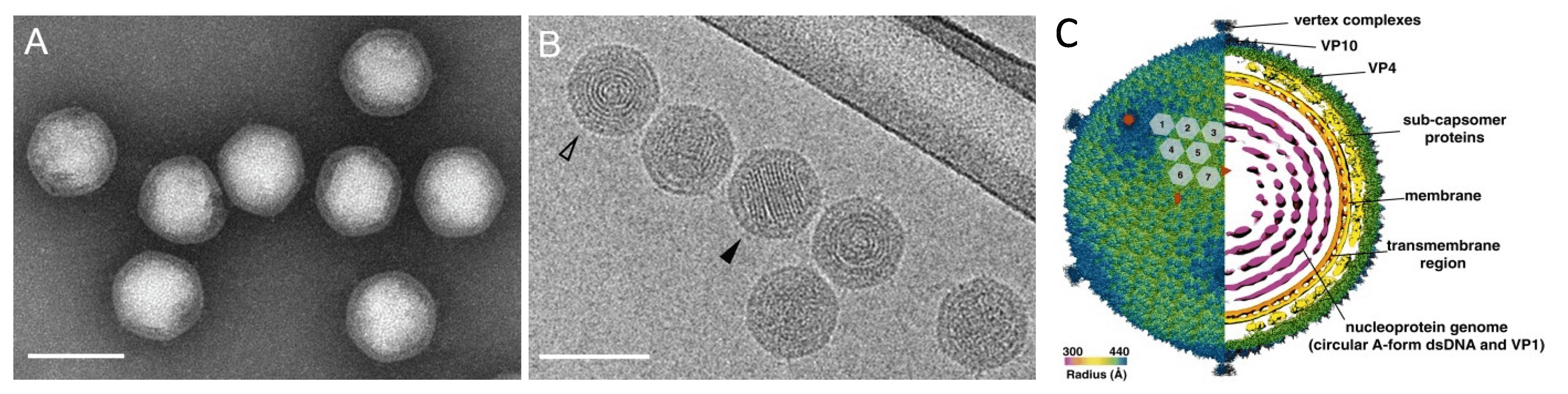 |
| Figure 1. . Portogloboviridae. Electron micrographs of Sulfolobus polyhedral virus 1 (SPV1) virions. (A) Negatively-stained with 2% (wt/vol) uranyl acetate. (B) Sample embedded in vitreous ice. The open arrowhead points to the projection in the axial view, the filled arrowhead points to the projection in the side view. Scale bars, 100 nm. (A, B from (Liu et al., 2017) Copyright © American Society for Microbiology) (C) Overall cryo-EM density map of SPV1, rendered to display the outer capsid (left half) and a central slice showing the interior of the virus (right half). The virus is colored by radius. The red rectangle, triangle and pentagon denote the two-fold, three-fold and five-fold axes of symmetry, respectively. The grey numbered hexagons outline the hexameric VP4 capsomers from one asymmetric unit of the SPV1 icosahedral capsid (from (Wang et al., 2019)). |
Physicochemical and physical properties
The outer protein shell can be removed from the intact virions by one cycle of freezing and thawing or prolonged storage. Subsequent treatment of the remaining cores with non-ionic detergents leads to partial disintegration of the lipid-containing membrane and the exposure of the globular nucleocapsid, which further unwinds into a nucleoprotein filament (Liu et al., 2017).
Nucleic acid
The circular dsDNA genomes of Sulfolobus polyhedral virus 1 (SPV1) and Sulfolobus polyhedral virus 2 (SPV2) are 20,222 bp and 20,424 bp long, respectively, and have a GC content of 38.3%, which is slightly higher than that of the host, Sacharolobus shibatae (35.5%) (Liu et al., 2017, Liu et al., 2019). The major nucleocapsid protein tightly wraps around the viral dsDNA, transforming it into the A-form (Wang et al., 2019).
Proteins
The virion contains four major proteins (VP1–VP4) and six minor proteins (VP5–VP10) (Liu et al., 2017). None of the major virion proteins show similarity to structural proteins of known viruses (Krupovic et al., 2018). However, structural studies have shown that the major capsid protein VP4 and the minor capsid protein VP10 which are present in the capsid at a ratio of 42:1, both have the jelly-roll fold (Wang et al., 2019), commonly found in icosahedral RNA and DNA viruses (Krupovic and Koonin 2017). VP1 is the major nucleocapsid protein and contains 4 α-helices of similar lengths (Wang et al., 2019). VP3, VP5, VP6, and VP7 have transmembrane helices and most likely reside in the viral membrane.
Lipids
Lipids are selectively acquired from the pool of host lipids and are integral constituents of the virion. The composition of the lipid species in SPV1 virions is quantitatively very different from that in the host. The host membrane is dominated (97%) by membrane-spanning C40 glycerol dibiphytanyl glycerol tetraether (GDGT) species, especially GDGT-3, GDGT-4, GDGT-5 and GDGT-6, whereas the virion membrane is specifically enriched in archaeol (42%) and GDGT-0 (20%), which together represent less than 2% of lipids in the host membrane (Wang et al., 2019).
Carbohydrates
No information available.
Genome organization and replication
The genomes of SPV1 and SPV2 contain 45 and 46 open reading frames (ORFs), respectively. The two viruses display an overall 91.7% pairwise genome sequence identity and are classified into two different species (Liu et al., 2017, Liu et al., 2019). The differences between the genomes are mainly caused by rearrangements, including insertions, deletions and one duplication. The ORFs are generally short, with a median of 103 codons, and are tightly arranged, occupying 89.1% of the genome (Figure 2. Portogloboviridae). Ten ORFs have homologs in various viruses infecting hosts of the order Sulfolobales, including members of the families Rudiviridae, Lipothrixviridae, Fuselloviridae and Turriviridae. Sequence analyses allowed the assignment of putative functions to nine ORFs, seven of which encode putative proteins containing various DNA-binding domains, including zinc finger, helix-turn-helix and ribbon-helix-helix domains. Other encoded proteins with predicted functions include the glycosyltransferase and the S-adenosyl L-methionine-dependent methyltransferase (Figure 2. Portogloboviridae) (Liu et al., 2017).
The viruses do not encode identifiable DNA and RNA polymerases or other known replication initiation proteins and, as in the case of most other viruses that infect the Sulfolobales, apparently depend on the host machinery for genome replication and transcription (Prangishvili et al., 2017).
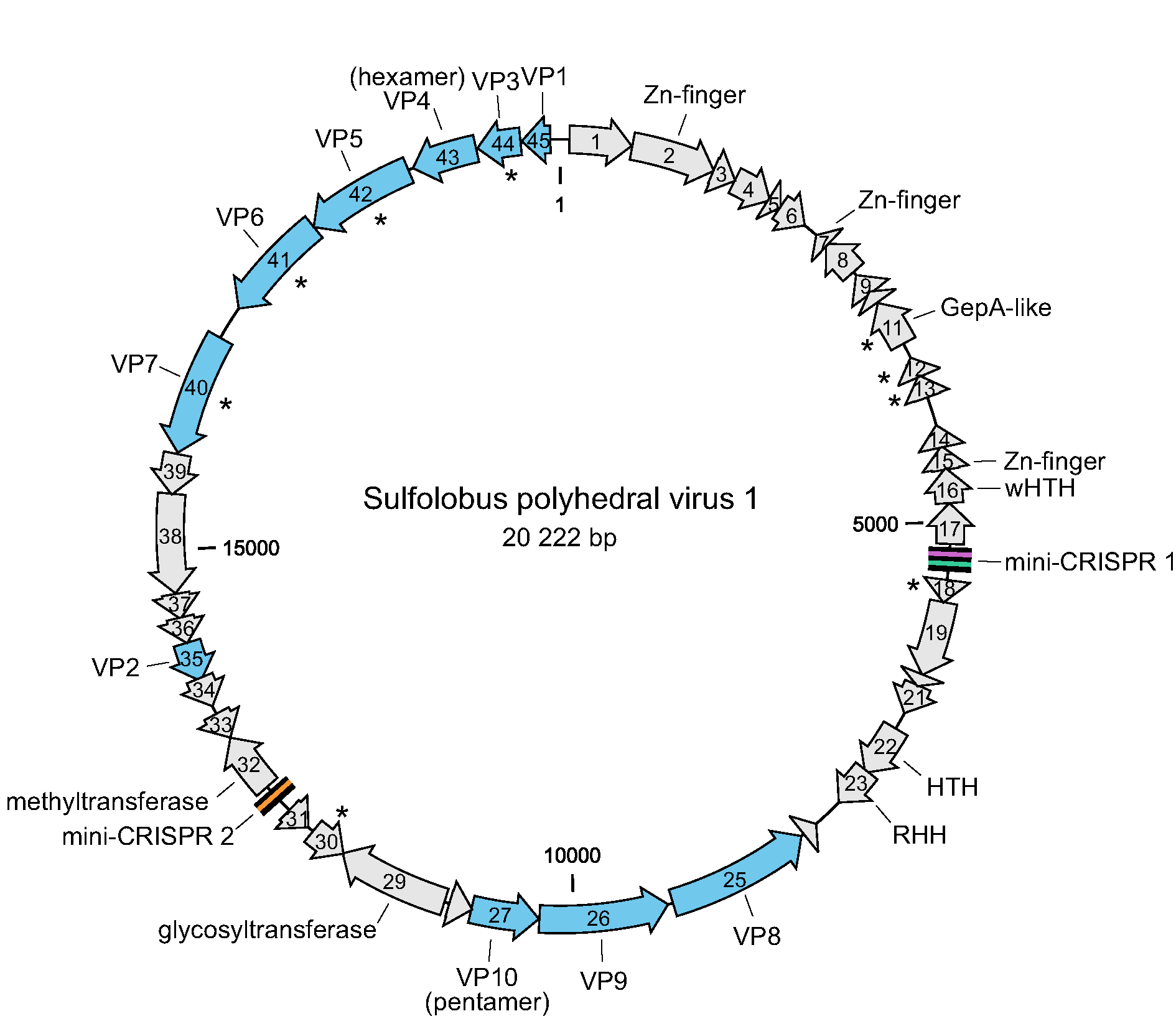 |
| Figure 2. Portogloboviridae. Genome map of Sulfolobus polyhedral virus 1. The open reading frames are represented with arrows that indicate the direction of transcription. Genes encoding virion structural proteins are shown in blue; those encoding predicted membrane proteins are indicated with asterisks. Abbreviations: (w)HTH, (winged) helix-turn-helix; RHH, ribbon-helix-helix. The locations of two mini-CRISPR arrays are indicated. |
Biology
The viruses SPV1 and SPV2 were identified in different environmental samples collected from two hot springs (75–80 °C, pH 3) in Beppu, Japan (Liu et al., 2017, Liu et al., 2019). The host range is limited to hyperthermophilic archaea from the genus Saccharolobus (order Sulfolobales). Viral replication occurs by chronic infection without an apparent lytic cycle. No information is available on the entry or egress of portogloboviruses. Portogloboviruses SPV1 and SPV2 carry mini-CRISPR arrays containing spacers targeting each other as well as other viruses, exemplifying a mechanism promoting inter-viral conflicts and superinfection exclusion in extreme environments (Medvedeva et al., 2019).
Derivation of names
Portogloboviridae: from the Latin words portare for ‘to carry’ and globus for ‘a ball’, in reference to the round shape of the nucleoprotein core.
Relationships within the family
Only two viruses have been described, which are 91.7% identical in genome sequence and classified into two different species (Liu et al., 2017, Liu et al., 2019).
Relationships with other taxa
Ten ORFs have homologs in various viruses infecting hosts of the order Sulfolobales, including members of the families Rudiviridae, Lipothrixviridae, Fuselloviridae and Turriviridae.

