Family: Quadriviridae
Sotaro Chiba, José R. Castón, Said A. Ghabrial and Nobuhiro Suzuki
The citation for this ICTV Report chapter is the summary published as Chiba et al., (2018):
ICTV Virus Taxonomy Profile: Quadriviridae, Journal of General Virology, 99, 1480–481
Corresponding author: Sotaro Chiba ([email protected])
Edited by: Peter Simmonds and Stuart G. Siddell
Posted: September 2018, updated May 2024
PDF: ICTV_Quadriviridae.pdf
Summary
The Quadriviridae is a family of non-enveloped spherical viruses with quadripartite dsRNA genomes of 3.5–5.0 kbp, comprising 16.8–17.1 kbp in total (Table 1 Quadriviridae). The family includes the genus Quadrivirus that includes the species Quadrivirus ichi. All quadriviruses have been reported to infect filamentous fungi. Pathogenicity of the viruses has not been reported. Quadriviruses have unique features in virion structure compared to those of other known dsRNA viruses. Quadriviruses are transmitted intracellularly via anastomosis of compatible fungal hyphae. Neither vertical transmission of the viruses through sexual and asexual spores or by natural vectors has been reported, and nor is it known if viruses have an extracellular phase.
Table 1 Quadriviridae. Characteristics of members of the family Quadriviridae.
| Characteristic | Description |
| Typical member | Rosellinia necatrix quadrivirus 1-W1075, (RNA1: AB620061; RNA2: AB620062; RNA3: AB620063; RNA4: AB620064), species Quadrivirus ichi, genus Quadrivirus |
| Genome | Four, linear dsRNAs of 3.5–5.0 kbp, 16.8–17.1 kbp in total |
| Virion | Isometric, non-enveloped, 45 nm in diameter; dsRNA segments may be separately encapsidated |
| Replication | No information available |
| Translation | Presumed from monocistronic positive-sense transcripts of each genomic dsRNA |
| Host Range | Fungi |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Duplornaviricota, class Chrymotiviricetes, order Ghabrivirales; one genus including three species |
Virion
Morphology
Quadriviruses form rigid spherical particles of 45 nm in diameter (Figure 1 Quadriviridae). Each capsid consists of 60 copies of heterodimers of two major structural proteins P2 and P4 encoded by dsRNA2 and dsRNA4, respectively (Figure 2 Quadriviridae). The P2 and P4 proteins form an asymmetric hetero-dimer subunit, and 60 of those build the T=1 capsid (Luque et al., 2016). The P2 and P4 proteins of the Rosellinia necatrix quadrivirus 1 (RnQV1) strains W1075 and W1118 are more than 80% identical. Despite a lack of sequence similarity between the two proteins, they have a similar a-helical domain, the structural signature shared with the lineage of the dsRNA viruses. While P2-P4 heterodimers are organized similarly to homodimers of other spherical dsRNA viruses, P2 and P4 have acquired domains situated on the outer surface that are hypothesized to possess actin regulatory (P2-domain) and protease (P4-domain) enzyme activities (Mata et al., 2017). Transfection competency of purified virions has not been demonstrated.
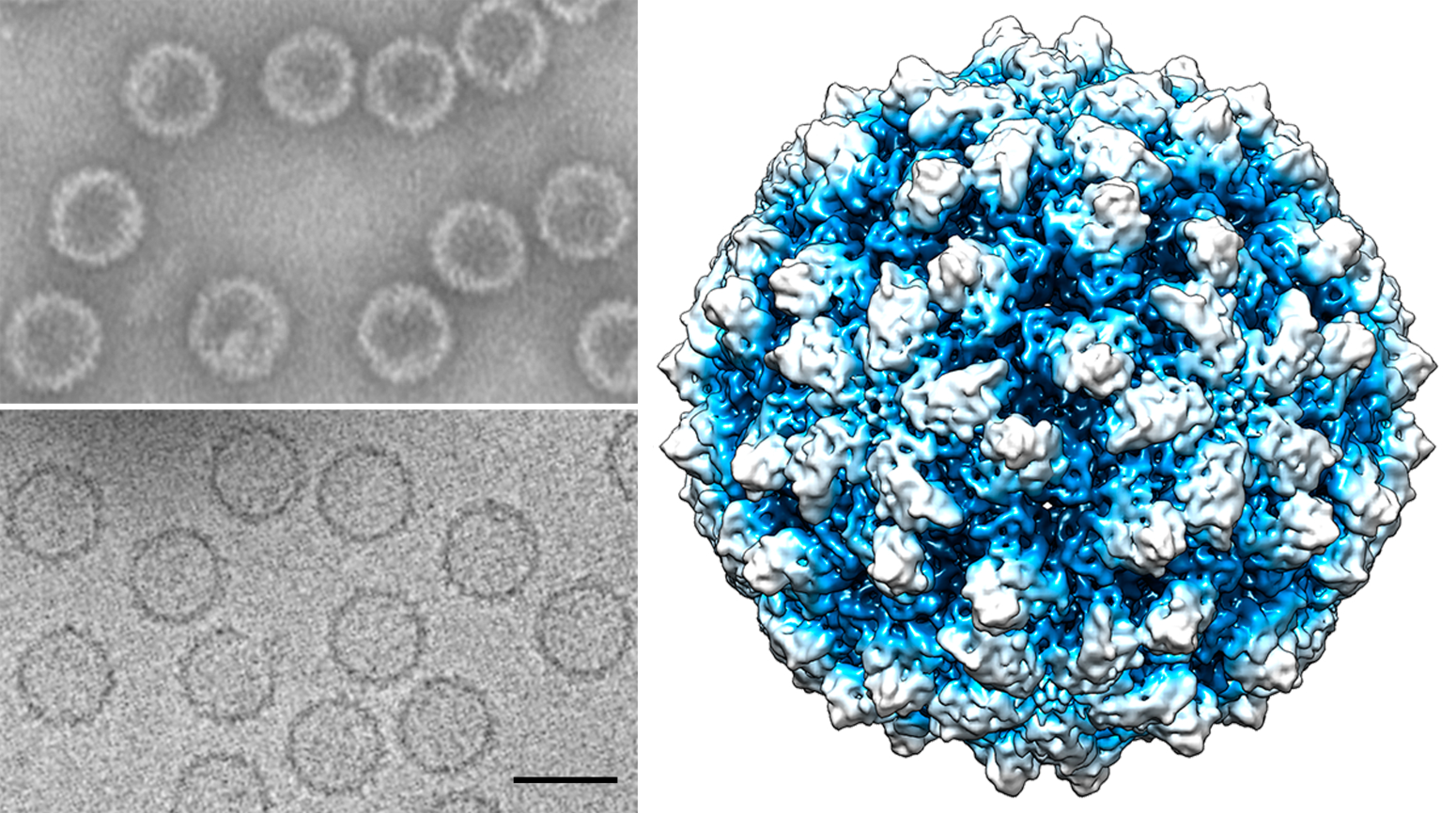 |
| Figure 1 Quadriviridae. Transmission electron micrograph of negatively stained (top, left) and unstained and vitrified (bottom, left) virions of the W1075 isolate of Rosellinia necatrix quadrivirus 1, a member of the species Quadrivirus ichi in the family Quadriviridae. Bar, 50 nm. (Right) Three-dimensional cryo-electron microscopy reconstruction of RnQV1 virions at 8Å resolution. |
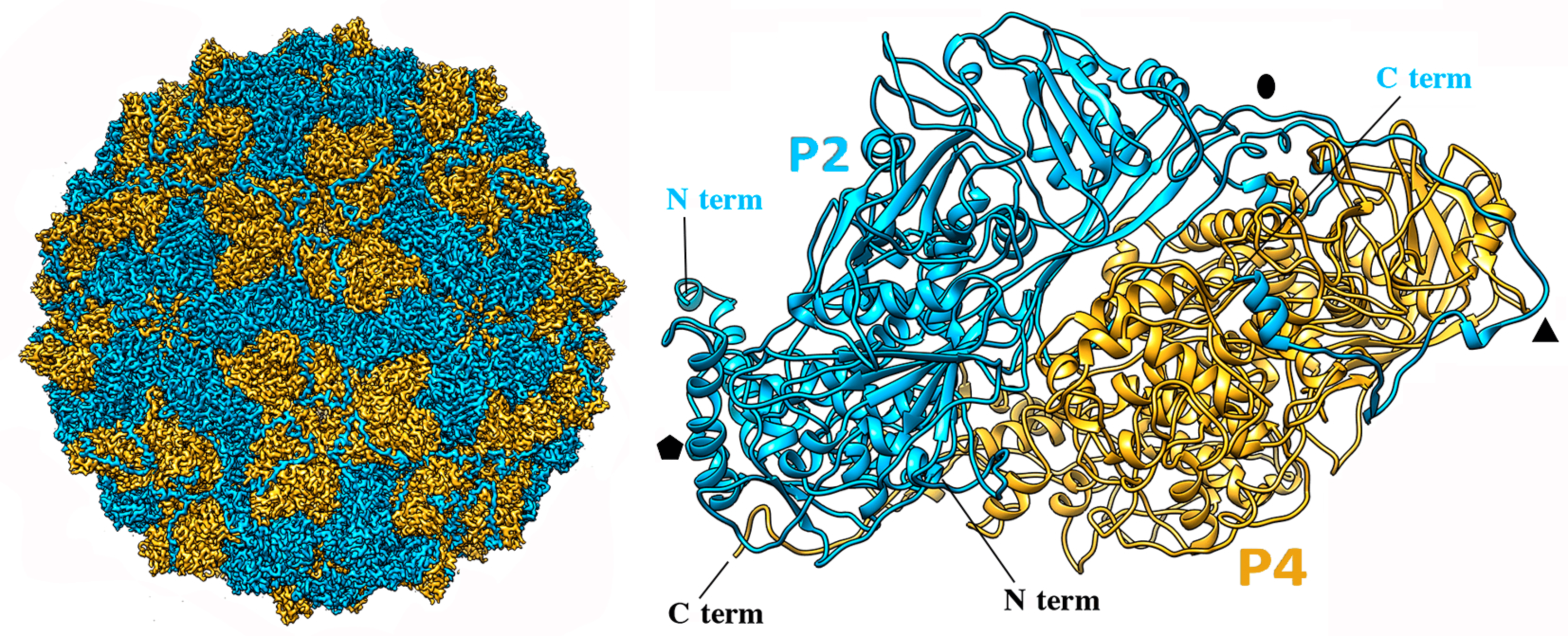 |
| Figure 2 Quadriviridae. Three-dimensional cryo-EM reconstruction of Rosellinia necatrix quadrivirus 1 virions (W1118 isolate) at a resolution of 3.7 Å. (Left) Cryo-EM map of the Rosellinia necatrix quadrivirus 1 capsid viewed along a twofold axis, showing P2 (blue) and P4 (yellow) proteins. (Right) Atomic model of P2 (blue) and P4 (yellow) (top view). N and C termini are indicated. Symbols indicate icosahedral symmetry axes; from (Mata et al., 2017). |
Physicochemical and physical properties
Unknown.
Nucleic acid
All members of the family Quadriviridae possess four dsRNA genome segments, termed dsRNA1 to dsRNA4 in a decreasing order of length. A single large ORF is present on the positive-sense strand of each of the dsRNA genomic segments (Figure 3 Quadriviridae). The linear dsRNA segments are 3.5-5.0 kbp and are expected to be encapsidated separately (Lin et al., 2012, Lin et al., 2013). The 5′-terminal 7 and 3′-terminal 13 nucleotides for RnQV1-W1075 and the 5′-terminal 9 and 3′-terminal 12 nucleotides for RnQV1-W1075, are conserved in all the genomic segments. There is notable heterogeneity in the very terminal ends of each segment (5′-C/U--------------G/A-3′) (Lin et al., 2012, Lin et al., 2013).
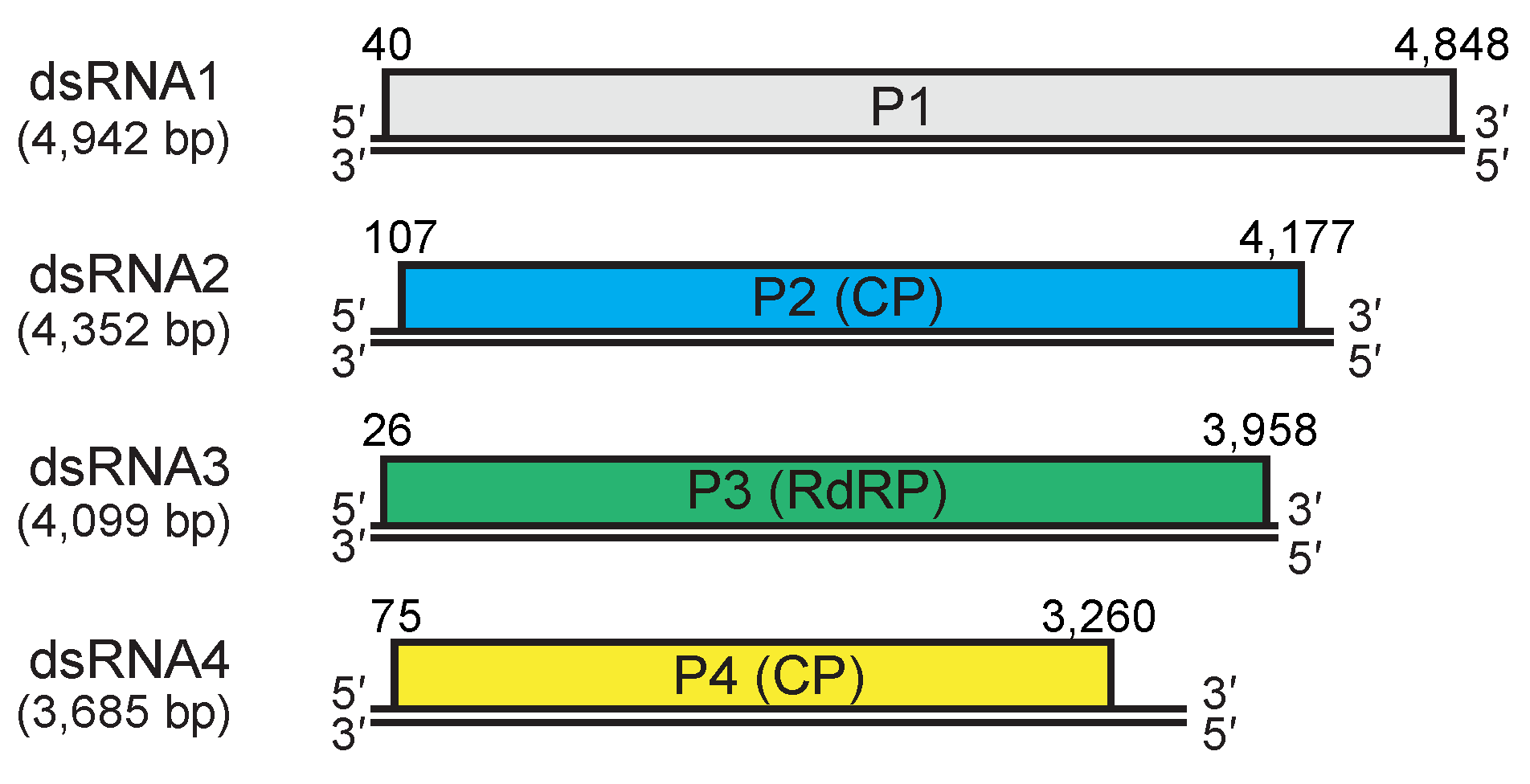 |
| Figure 3. Quadriviridae. Genomic organization of the W1075 strain of Rosellinia necatrix quadrivirus 1. dsRNA2 and dsRNA4 encode structural proteins (capsid protein, CP) while dsRNA3 encodes the replicase (RNA-dependent RNA polymerase, RdRP). dsRNA1 encodes a non-structural protein of unknown function. Double lines indicate genomic dsRNAs; boxes indicate open reading frames on the positive-sense strands which begin and end at the nucleotide positions indicated. |
Genome organization and replication
Quadrivirus genomes consist of four linear, monocistronic dsRNA segments; the largest segment/ORF (dsRNA1) encodes a hypothetical protein (P1) with unknown function. The second and fourth segments (dsRNA2 and dsRNA4) encode the capsid proteins (CP, P2 and P4 respectively), whereas the third segment (dsRNA3) encodes viral RdRP (Lin et al., 2012, Lin et al., 2013) (Figure 3 Quadriviridae). Both the 5′- and 3′- terminal sequences of the dsRNA genome are well conserved in all the segments. Transcription and replication are presumably carried out in a spherical particle by virion-associated RdRP. All the genomic segments except for dsRNA3 have (CAA)n repeats, which are presumably translational enhancers, as reported for chrysoviruses (Ghabrial et al., 2018) and some partitiviruses (Chiba et al., 2016, Tavantzis 2008).
Biology
Quadriviruses persistently infect their fungal hosts, and are stably maintained in the natural host Rosellinia necatrix (an ascomycete fungus) for a long period, despite successive subcultures. Quadriviruses are transmitted intracellularly during cell division and hyphal anastomosis (Lin et al., 2012). Horizontal transmission by hyphal fusion between fungi of different mycelial compatibility groups is blocked by programmed cell death (PCD). PCD appears to be retarded by the presence of Zn2+ cations in the media, which allows mycoviruses to be horizontally transmitted between vegetatively incompatible mycelia, as is the case for a megabirnavirus (Rosellinia necatrix megabirnavirus 1) (Ikeda et al., 2013, Kondo et al., 2013). Little is known about vertical transmission during asexual or sexual sporulation, or horizontal transmission by natural vectors. The quadrivirus RnQV1-W1075 spreads relatively slowly and shows uneven distribution in fungal colonies compared to other R. necatrix-infecting viruses (Yaegashi et al., 2011). Attempts to transfect protoplasts derived from the natural (R. necatrix) and experimental fungal hosts (Cryphonectria parasitica) with purified quadriviruses have been unsuccessful.
Antigenicity
Antibodies prepared to N- and C-terminal truncated polypeptides derived from RnQV1-W1075 P2 show cross-reactivities to RnQV-W1118 P2 (Lin et al., 2013).
Derivation of names
Quadriviridae, Quadrivirus: from the viruses’ quadripartite genome, from the Latin quad meaning four.
Relationships within the family
Three strains of RnQV1 have been partially or completely sequenced and compared. The exemplar virus, RnQV1 W1075 shows greater phylogenetic affinity to RnQV1 W726 than to W1118. Greater sequence similarity is observed between strains W1075 and W726 than between other combinations. Amino acid sequence deduced from the W726 dsRNA3 shows 98% and 75% identities to the corresponding portions of W1075 and W118 RdRPs, respectively. Amino acid sequence identities found between the homologues of RnQV1 W1075 and W1118 vary from 72% (P1) to 82% (P4), while those for nucleotide sequence identities vary from 67% (dsRNA1 and dsRNA3) to 70% (dsRNA2).
Relationships with other taxa
Phylogenetic analyses show that quadriviruses are most closely related to members of the genus Totivirus in the family Totiviridae (Lin et al., 2012, Lin et al., 2013) (Figure 4 Quadriviridae). Other quadripartite dsRNA viruses such as chrysoviruses and unclassified alternaviruses show relatively distant relation to quadriviruses. It is noteworthy that members of the genus Totivirus show greater phylogenetic affinity to quadriviruses than to members of other genera within Totiviridae (Figure 4 Quadriviridae). This suggests that taxonomic reorganization of the family Totiviridae needs to be considered.
The quadripartite nature of quadrivirus dsRNA genomes resembles that of members of the family Chrysoviridae and an unclassified mycovirus Alternaria alternata virus 1 (AaV-1). However, the size range of genomic dsRNA segments are different for each of these viruses: quadriviruses, 3.5–5.0 kbp; chrysoviruses, 2.5–3.6 kbp (Ghabrial et al., 2018); AaV-1, 1.4–3.6 kbp excluding a poly(A) tract (Aoki et al., 2009). The 5′-UTRs of quadriviruses and AaV-1 are short (24–107 bp), and those of chrysoviruses are relatively long, presumably due to the possession of an internal ribosomal entry site (Chiba et al., 2018). The 3′-end of AaV-1 genomic segments are polyadenylated in the coding strand, similar to some members in the family Partitiviridae, which is not found in quadriviruses and chrysoviruses (Lin et al., 2012, Aoki et al., 2009). Chrysoviruses and AaV-1 form a capsid with a single CP, whereas quadriviruses have two CPs. A taxonomically unclassified quadripartite dsRNA virus (1.1–2.4 kbp) tentatively named tetramycovirus was reported (Kanhayuwa et al., 2015) and recently renamed as a polymycovirus upon the discoveries of similar viruses with more than 4 genomic segments (Kotta-Loizou and Coutts 2017). Although the polymycovirus genome segments are monocistronic, as are the genome segments of the abovementioned quadripartite dsRNA viruses, polymycoviruses do not form typical virions and are infectious as purified viral dsRNA (Kanhayuwa et al., 2015, Kotta-Loizou and Coutts 2017).
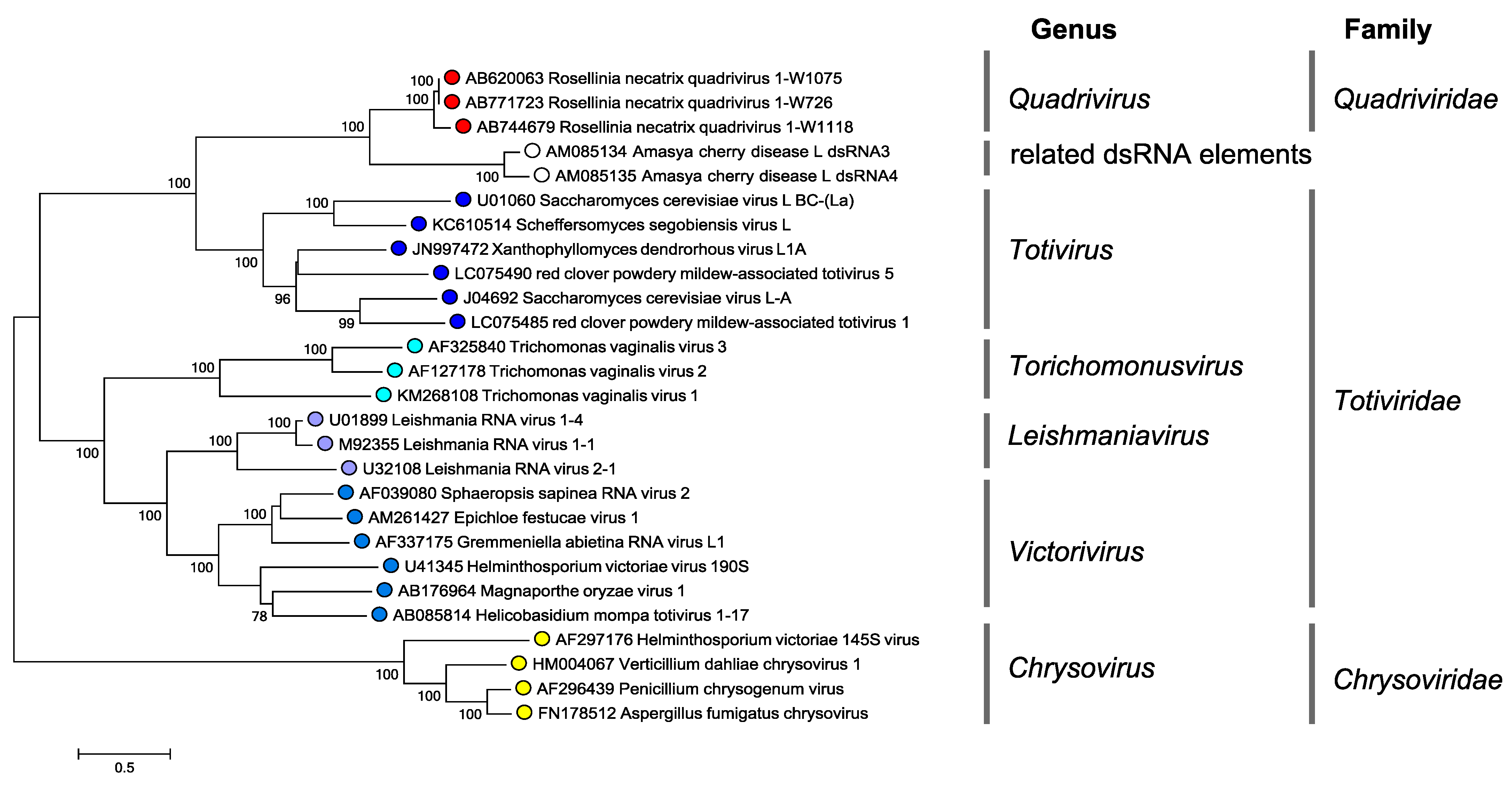 |
| Figure 4 Quadriviridae. Phylogenetic tree of the RdRP of quadriviruses, chrysoviruses and some members of the family Totiviridae. Translated sequences were aligned using MUSCLE (Edgar 2004) as implemented in SSE v1.3 (Simmonds 2012), and the least divergent region (amino acid positions 679 to 1066, numbered according to the RdRP of Rosellinia necatrix quadrivirus 1 strain W1075) was used to produce a midpoint rooted maximum likelihood dendrogram in MEGA7 (Kumar et al., 2016) using the LG model with a gamma distribution of substitution rates and a proportion of invariant sites. Numbers at the nodes denote bootstrap values where these were > 70%. The scale bar represents the amino acid distances. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Related, unclassified viruses
Virus-like dsRNA elements with high sequence similarity to RnQV1 dsRNA1 and dsRNA3 have been reported (Lin et al., 2012, Kozlakidis et al., 2006) (Figure 4 Quadriviridae). These elements are named after Amasya cherry disease (ACD)-dsRNA1, -dsRNA2, -dsRNA3 and -dsRNA4, and occur together with a chrysovirus and a partitivirus (Coutts et al., 2004, Covelli et al., 2004). However, no CP-encoding dsRNA segments (corresponding to RnQV1 dsRNA2 and dsRNA4) have been reported. Such dsRNAs were also found in cherry chlorotic rusty spot (CCRS)-associated cherry leaves (Kozlakidis et al., 2006). ACD and CCRS are diseases presumably with a fungal etiology, but isolation of the causal pathogens has not been successful. This leaves unanswered the question of whether the origin of these dsRNA elements is from fungal or plant tissue.
| Probable viral element | RnQV1-dsRNA1 like | RnQV1-dsRNA3 like | Virus abbreviation |
| Amasya cherry disease L dsRNA | dsRNA1: AM085136; dsRNA2: AM085137 | dsRNA3: AM085134; dsRNA4: AM085135 | ACD-L |
Virus names and virus abbreviations are not official ICTV designations.

