Family: Secoviridae
Marc Fuchs, Jean-Michel Hily, Karel Petrzik, Hélène Sanfaçon, Jeremy R. Thompson, René van der Vlugt and Thierry Wetzel
The citation for this ICTV Report chapter is the summary published as Fuchs M, Hily JM, Petrzik K, Sanfaçon H, Thompson JR, van der Vlugt R, Wetzel T. and ICTV Report Consortium (2022):
ICTV Virus Taxonomy Profile: Secoviridae 2022, Journal of General Virology (2022) 103:001807
Corresponding author: Marc Fuchs ([email protected])
Edited by: Luisa Rubino
Posted: April 2017, updated September 2019, October 2022, May 2023, July 2025
Summary
Members of the family Secoviridae are non-enveloped viruses with mono- or bipartite (RNA1 and RNA2) linear positive-sense ssRNA genomes of 9 to 13.7 kilobases in total (Thompson 2020). Secoviruses are related to picornaviruses and are classified in the order Picornavirales. The majority of known members infect dicotyledonous plants, and many are important plant pathogens (e.g. grapevine fanleaf virus and rice tungro spherical virus) (Sanfaçon 2015).
Table 1 Secoviridae. Characteristics of members of the family Secoviridae
| Characteristic | Description |
| Example | cowpea mosaic virus (RNA1: X00206; RNA2: X00729), species Comovirus vignae, genus Comovirus |
| Virion | Non-enveloped 25–30 nm in diameter with icosahedral symmetry |
Genome | 9 to 13.7 kb of positive-sense, mono- or bipartite RNA |
Replication | In association with intracellular membranes derived from the endoplasmic reticulum |
| Translation | Directly from genomic RNA as large polyproteins, which are cleaved by RNA1-encoded 3C-like proteinases in cis and trans, and, for stramoviruses, also by an RNA2-encoded glutamic protease in cis |
| Host range | Plants (mainly dicots), transmitted mainly by insects or nematodes. Some seed transmission demonstrated |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Pisuviricota, class Pisoniviricetes, order Picornavirales; the family includes one sub-family with four genera, six additional genera, three sub-genera, and 165 species |
Comovirus - (subfamily Comovirinae). Bipartite genome. The genus includes 19 species. Comoviruses usually have a narrow host range. Mosaic and mottle symptoms are characteristic. Transmission in nature is exclusively by beetles, especially members of the family Chrysomelidae. Beetles retain their ability to transmit virus for days or weeks.
Fabavirus - (subfamily Comovirinae). Bipartite genome. The genus includes eight species. Fabaviruses a wide host range among dicotyledonous and some families of monocotyledonous plants. Symptoms are typically ringspots, mottling and wilting. In nature, they are transmitted by aphids in a non-persistent manner.
Mersevirus - (subfamily Comovirinae). Bipartite genome. The genus includes four species. A Ham1 domain with predicted inosine triphosphate pyrophosphatase activity is present at the C-terminus of the RNA-directed RNA polymerase. The pathogenicity of merseviruses is unclear and no information is available on their transmission.
Nepovirus - (subfamily Comovirinae). Bipartite genome. The genus includes 48 species. Nepoviruses are widely distributed in temperate regions. Ringspot symptoms are characteristic. Many nepoviruses are transmitted non-persistently by longidorid nematodes. Seed and/or pollen transmission are also common. In herbaceous plants, the symptoms induced are often transient with a so-called “recovery” phenomenon.
Cheravirus. Bipartite genome. The genus includes five species. Cheraviruses usually cause mild symptoms or asymptomatic infections. Cherry rasp leaf virus is transmitted by nematodes in the field.
Sadwavirus. Bipartite genone. The genus includes nine species in three sub-genera (Cholivirus, Satsumavirus and Stramovirus). The host range is narrow, and the geographic distribution is usually restricted. The natural mode of transmission is unknown.
Sequivirus. Monopartite genome. The genus includes four species. The natural host range of sequiviruses includes plants in several families. Transmission is by aphids in a semi-persistent manner, but is dependent on the presence of a helper virus in the genus Waikavirus.
Stralarivirus. Bipartite genome. The genus includes two species. The host range of stralariviruses includes plants in several families and the geographic distribution is broad. Strawberry latent ringspot virus is transmitted by nematodes.
Torradovirus. Bipartite genome. The genus includes eight species. The torradovirus genome contains an open reading frame (ORF) upstream and partially overlapping the large ORF2 in RNA2. Some torradoviruses are transmitted by whiteflies in a semi-persistent manner; one torradovirus is transmitted by aphids.
Waikavirus. Monopartite genome. The genus includes 12 species. The natural host range of waikaviruses is usually restricted to plants within a few families. Field transmission is semi-persistent by aphids or leafhoppers. Some waikaviruses are helper viruses for the insect transmission of other viruses. For example, rice tungro spherical virus is the helper virus for rice tungro bacilliform virus (family Caulimoviridae).
Virion
Morphology
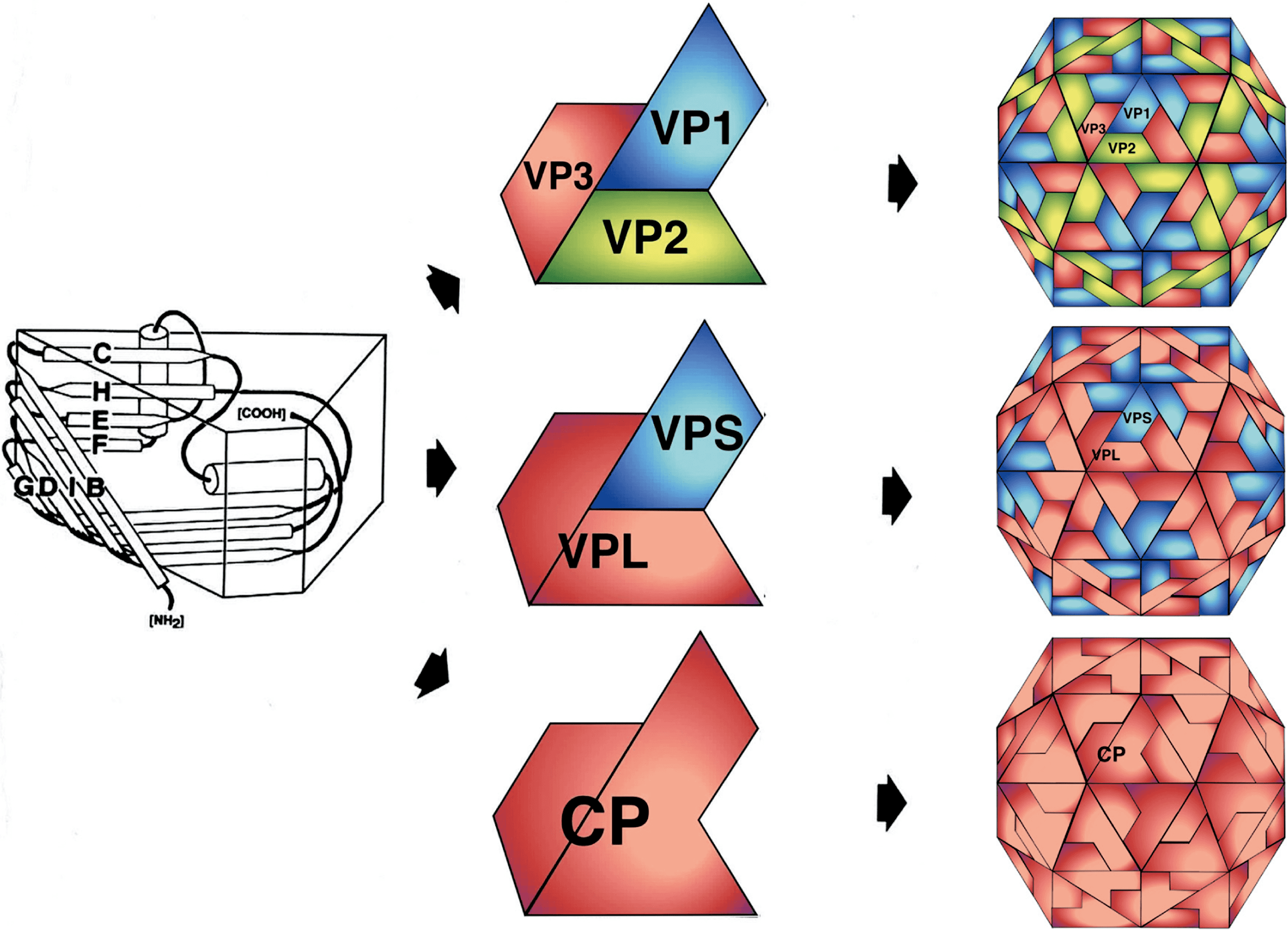
Virions are non-enveloped 25–30 nm in diameter and exhibit icosahedral symmetry (T=1, pseudo T=3, Figure 1 Secoviridae) (Lin and Johnson 2003). Many virus preparations contain empty particles. In the case of viruses with a bipartite genome, the two RNAs are encapsidated in separate virions.
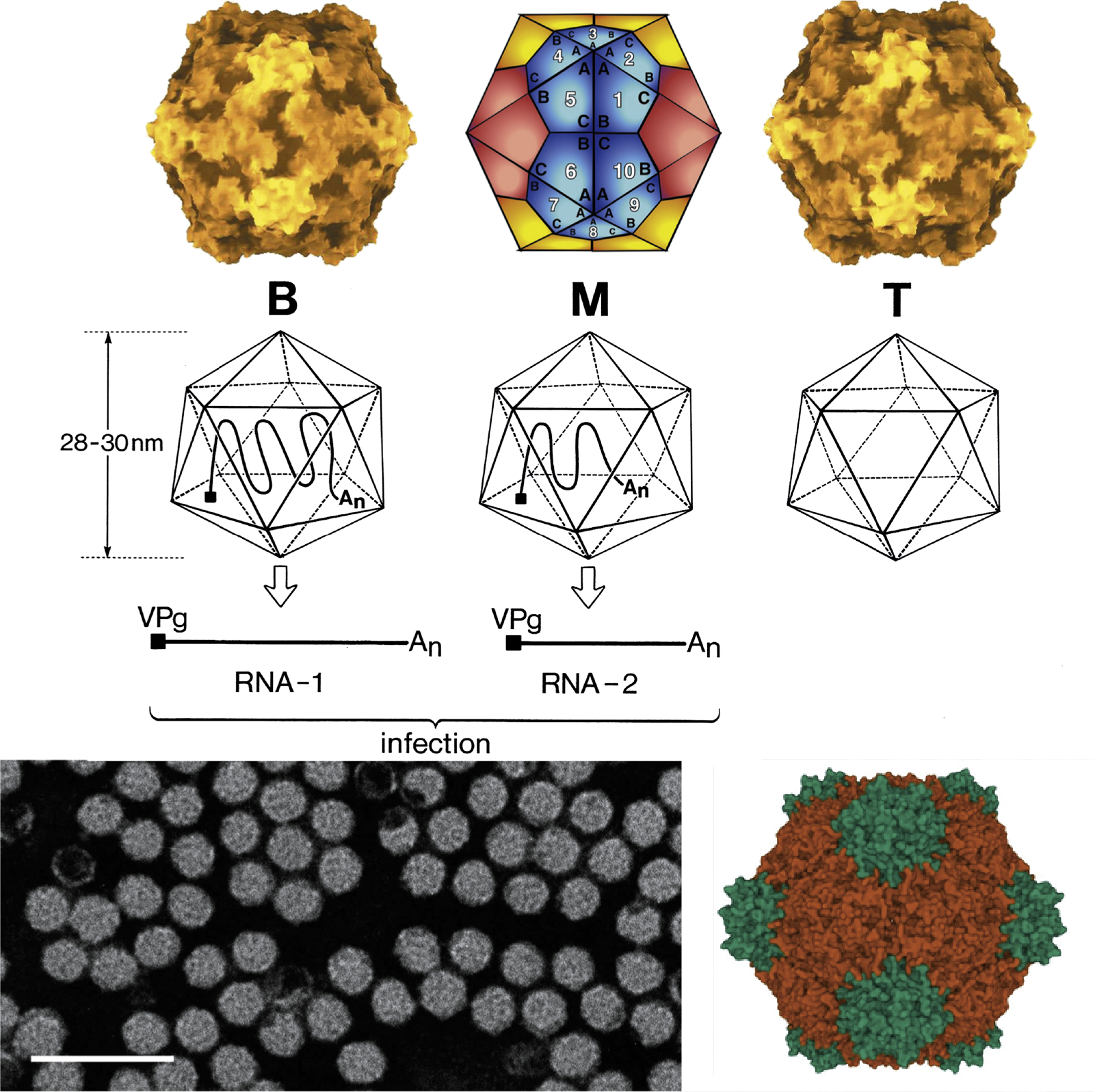 |
| Figure 1 Secoviridae. (Top left): Molecular rendering of the cowpea mosaic virus particle. (Top central): Diagrammatic representation of a T=1 lattice. A = Small capsid protein, B = C-terminal domain of the large capsid protein and C=N-terminal domain of the large capsid protein. (Top right): Molecular rendering of the red clover mottle virus particle. (Center): Diagram of the three types of comovirus particles with the B-particle containing one molecule of RNA1, the M-particle containing one molecule of RNA2 and the T-particle being empty. (Bottom left): Negative-contrast electron micrograph of particles of cowpea mosaic virus. The bar represents 100 nm. (Bottom right): Crystal structure of the capsid of cowpea mosaic virus with the S (pine green) and L (dark orange) coat protein subunits forming three ß-sandwich domains (PDB,10.2210/pdb1NY7/pdb). |
Physicochemical and physical properties
Different classes of virions are distinguished according to their buoyant densities (top, middle and bottom components, also termed T, M and B), (Figure 1 Secoviridae). The virion components M and B contain RNA. Viruses belonging to the genera Sequivirus and Waikavirus, which have a large monopartite genome, sediment with S20W values of 150–190S. For viruses with a bipartite genome, virions containing RNA1 (B component) sediment at 110–135S. Virions containing RNA2 (M component) sediment at 84–128S and contain one or two molecules of RNA2. In cases where the lengths of RNA1 and RNA2 are similar, the M and B components may be difficult to separate. Empty shells (T component) sediment with S20W values of 49–63S depending on the virus considered.
Nucleic acid
The genome consists of one or two molecules of linear positive-sense ssRNA with lengths that differ among genera (Table 2 Secoviridae). The genomic RNA(s) contain a 3ʹ-terminal poly(A) tract of variable length. The only known exception is the genomic RNA of a sequivirus (parsnip yellow fleck virus), which is apparently not polyadenylated. For several comoviruses and nepoviruses, and for strawberry latent ringspot virus, a protein, designated VPg (2–4 kDa) has been shown to be covalently bound at the 5ʹ-end. The presence of a 5ʹ-linked VPg has not been confirmed for other genera but has been suggested because in many cases, infectivity of the RNA(s) has been shown to be protease-sensitive.
Table 2 Secoviridae. Accession numbers and genome content (bases) of representative viruses in the family Secoviridae
| Subfamily/genus/virus | RNA1 | RNA2 |
|---|---|---|
| Comovirinae | ||
| Comovirus | 5,850–6,100 | 3,300–4,000 |
| cowpea mosaic virus-van Wezenbeek | X00206 (5,889) | X00729 (3,481) |
| Fabavirus | 5,800–6,000 | 3,300–4,000 |
| broad bean wilt virus 2-ME | AF225953 (5,951) | AF225954 (3,607) |
| Mersevirus | 6,000–6,300 | 3,200–3,550 |
| Mercurialis secovirus 1 | OR544055 | OR544056 |
| Nepovirus | 7,200–8,400 | 3,700–7,300 |
| grapevine fanleaf virus-F13 (Subgroup A) | D00915 (7,342) | X16907 (3,774) |
| beet ringspot virus-S (Subgroup B) | D00322 (7,356) | X04062 (4,662) |
| tomato ringspot virus-Raspberry (Subgroup C) | L19655 (8,214) | D12477 (7,273) |
| Not assigned to a subfamily | ||
| Cheravirus | 6,800–7,100 | 3,200–3,700 |
| cherry rasp leaf virus-USA | AJ621357 (6,992) | AJ621358 (3,274) |
| Sadwavirus | 6,800–7,000 | 5,300–5,600 |
| satsuma dwarf virus-S58 | AB009958 (6,795) | AB009959 (5,345) |
| Sequivirus | 9,800–10,000 | |
| parsnip yellow fleck virus-P121 | D14066 (9,871) | NA |
| Stralarivirus | 7,400–7,500 | 3,700–3,800 |
| strawberry latent ringspot virus-MEN 454 | AY860978 (7,496) | AY860979 (3,842) |
| Torradovirus | 7,000–7,800 | 4,700–5,900 |
| tomato torrado virus-PRI-0301 | DQ388879 (7,793) | DQ388880 (5,389) |
| Waikavirus | 11,700–12,500 | NA |
| rice tungro spherical virus-Shen | M95497 (12,226) |
NA: not applicable.
Proteins
Nepoviruses and some sadwaviruses (stramoviruses and choliviruses) have a single capsid protein (CP) of 52–60 kDa. Comoviruses, fabaviruses, some sadwaviruses (satsumaviruses) and stralariviruses have two CPs of 40–45 kDa and 21–29 kDa. Cheraviruses, torradoviruses, sequiviruses and waikaviruses have three CPs of similar sizes (24–35 kDa, 20–26 kDa and 20–25 kDa). For three comoviruses (cowpea mosaic virus, bean pod mottle virus and red clover mottle virus) and three nepoviruses (tobacco ringspot virus, grapevine fanleaf virus and arabis mosaic virus), the atomic structure has been solved and found to be very similar (pseudo T=3) to that of viruses belonging to the family Picornaviridae (Chen et al., 1989, Chandrasekar and Johnson 1998, Lin et al., 2000, Schellenberger et al., 2011, Lai-Kee-Him et al., 2013). Each capsid subunit consists of three beta-barrels (jelly roll domains) that can be present in one large CP with three jelly roll domains (Nepovirus and Sadwavirus), two CPs (one large CP including two jelly roll domains and one smaller CP with a single jelly roll domain; Comovirus, Fabavirus and Sadwavirus and Stralarivirus) or three CPs each containing a single jelly roll domain (Cheravirus, Torradovirus, Sequivirus, Waikavirus) (Figure 2 Secoviridae).
Genome organization and replication
Unfractionated viral RNA is highly infective. In the case of viruses with a bipartite genome, neither RNA species alone can infect plants systemically. RNA1 carries all the information required for replication and can replicate in individual cells in the absence of RNA2 although no virus particles are produced (as demonstrated for comoviruses and nepoviruses).
Viral proteins are usually expressed as large polyproteins, which are cleaved by RNA1-encoded 3C-like proteinases. An RNA2-encoded glutamic protease also cleaves the RNA2 polyprotein of stramoviruses (Mann et al., 2019). Each RNA usually encodes a single polyprotein (Figure 3 Secoviridae). A notable exception is the RNA2 of torradoviruses, which contains two ORFs. Another exception is the RNA2 of comoviruses. Although a single large ORF is present, internal initiation at a second AUG allows the formation of two distinct polyproteins. In some cases, extensive regions of sequence identity between RNA1 and RNA2 are found in the 5ʹ- and/or 3ʹ-untranslated regions (UTRs) (Figure 3.Secoviridae).
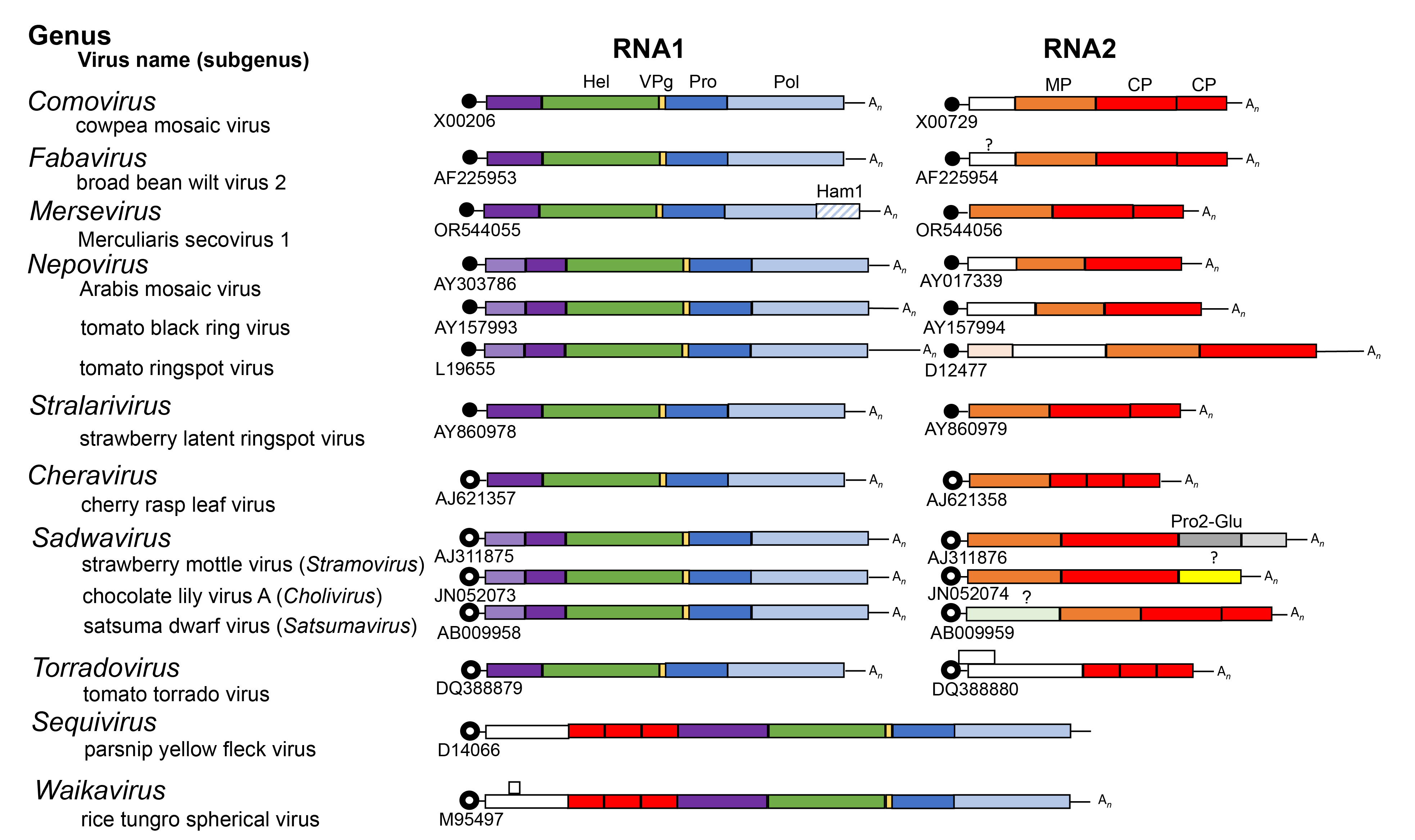 |
| Figure 3 Secoviridae. Genome organization of representative members of the 10 genera (Comovirus, Fabavirus, Mersevirus, Nepovirus, Stralarivirus, Cheravirus, Sadwavirus, Torradovirus, Sequivirus, Waikavirus) in the family Secoviridae. Each RNA is shown with open reading frames (ORFs) represented with boxes. Circles at the 5′-end of viral genomic RNA depict viral genome-linked proteins (VPg). Black circles represent VPg experimentally confirmed and open circles represent putative VPgs. The poly(A) tails at the 3′-end of viral genomic RNAs are depicted with (An), when appropriate. Colours indicate protein domains with conserved motifs for the putative NTP-binding domain protein (Hel, green), VPg (peach), 3C-like proteinase (Pro, dark blue), RNA-directed RNA polymerase (Pol, light blue), Ham1 domain (light blue diagonal stripes), movement protein (MP, orange) and coat protein(s) (CPs, red) are shown. Pro2-Glu (dark grey) is a glutamic protease of the sadwavirus strawberry mottle virus. Proteinase cleavage sites identified experimentally or predicted by sequence comparisons are indicated by solid vertical lines. The three sub-genera of sadwaviruses are indicated. |
Within the polyproteins, protein domains are organized in a manner common to that of other members of the order Picornavirales (Figure 3 Secoviridae). The replication block contains domains characteristic of NTP-binding proteins (NTB or putative helicase), 3C-like proteinase (Pro) and RNA-directed RNA polymerase (Pol). In viruses with a monopartite genome, the structural proteins are located upstream of the replication block in the single polyprotein. In viruses with a bipartite genome, structural proteins are contained in the RNA2-encoded polyprotein. In comoviruses, cheraviruses, nepoviruses and stralariviruses, the movement protein is located upstream of the CP(s), and enables viral movement to adjacent cells. Both movement protein and CP(s) are required for cell-to-cell movement of the virus. The movement protein of comoviruses and nepoviruses is a structural component of tubular structures that traverse the cell wall and contain virus-like particles (Laporte et al., 2003, Pouwels et al., 2004). Putative movement proteins have been suggested to be encoded upstream of the CP(s) coding regions for many other viruses in the family, but their biological function has not been confirmed.
The RNA1-encoded 3C proteinase cleaves both RNA1 and RNA2-encoded polyproteins. The cleavage site specificity of the proteinase differs with the specific genera (and in the case of nepoviruses it differs with the specific subgroup, Table 3 Secoviridae) (Wellink and van Kammen 1988, Gorbalenya et al., 1989, Margis and Pinck 1992, Thole and Hull 1998, Carrier et al., 1999, Ferriol et al., 2016). An amino acid in the substrate-binding pocket of the proteinase interacts directly with the amino acid in the −1 position of the cleavage site and plays a key role in the specificity of the proteinase (Sanfaçon 2022).
Table 3 Secoviridae. Cleavage site specificity of the 3C-like proteinase of viruses in the family Secoviridae
| Subfamily/ genus | Proteinase substrate binding pocket# | Dipeptide at cleavage site† |
|---|---|---|
| Comovirinae | ||
| Comovirus | His | Q/G, Q/M, Q/S, Q/T*, Q/A* |
| Fabavirus | His | Q/S*, Q/A*, Q/G* |
| Mersevirus | ? | Q/F, Q/H, Q/N, Q/F |
| Nepovirus | ||
| Subgroup A | Leu | R/G, C/S, C/A, A/S, G/E*, G/V*, C/G* |
| Subgroup B | Leu | K/S, K/A, R/A, R/S*, R/G* |
| Subgroup C | His | Q/G, Q/S, D/S |
| Not assigned to a subfamily | ||
| Cheravirus | His | Q/G, E/G |
| Sadwavirus◊ | ? | R/G, R/S, T/S, Q/G, Q/S, Q/A, E/G |
| Sequivirus | Leu | ? |
| Stralarivirus | ? | S/G |
| Torradovirus | His | Q/A, Q/S, Q/V |
| Waikavirus | His | Q/S*, Q/M*, Q/V*, Q/A* |
#The indicated amino acid in the substrate-binding pocket of the proteinase interacts with the amino acid at the −1 position of the cleavage site and plays an important role in determining the cleavage site specificity of the proteinase. In the case of sadwaviruses, neither a His nor a Leu are found at the equivalent position in the deduced amino acid sequence of the proteinase.
†Cleaved dipeptides at the cleavage sites are shown with the scissile bond indicated with the slanted line. The amino acids are shown using the one-letter code. Proteolytic cleavages at dipeptides have been confirmed experimentally except for those indicated by an asterisk which are putative cleavage sites, inferred from sequence alignments.
◊In the genus Sadwavirus, stramoviruses encode also a glutamic protease that cleaves the RNA2 polyprotein at P/AFP, PKFP, ALMP and PMYP site (Sanfaçon 2022).
Formation of replication complexes has been studied for comoviruses and nepoviruses. Replication occurs in association with intracellular membranes derived from the endoplasmic reticulum. Two RNA1-encoded proteins (the NTB protein and the protein immediately upstream of NTB) interact directly with ER membranes and have been implicated in the proliferation of membrane vesicles in the cytoplasm of infected cells and in the assembly of the replication complex (Sanfaçon 2012). This has not been studied for other viruses in the family.
Biology
All members of the family infect plants. Host range and symptoms vary with the genera and viruses considered (Table 4 Secoviridae). Many viruses in the family have a known biological vector, although sequiviruses require a helper virus and others do not have a known vector. Most viruses are transmissible experimentally by mechanical inoculation. However, waikaviruses are not known to be sap transmissible. Many viruses are readily transmissible by seed or pollen.
Table 4 Secoviridae. Biological properties of viruses in the family Secoviridae
| Subfamily/genus | Host range | Vector | Seed or pollen transmission |
|---|---|---|---|
| Comovirinae | |||
| Comovirus | Narrow (Leguminosae) | Beetle | Rare |
| Fabavirus | Wide | Aphid | Rare |
| Mersevirus | Relatively wide | Unknown | Unknown |
| Nepovirus | Wide | Nematode (most), mite (blackcurrant reversion virus) or unknown | Seed, pollen |
| Not assigned to a subfamily | |||
| Cheravirus | Wide or narrow | Nematode (cherry rasp leaf virus) or unknown | Seed |
| Sadwavirus | Wide | Aphid or unknown | Seed |
| Sequivirus | Relatively wide | Aphid (requires helper virus) | None |
| Stralarivirus | Wide | Nematode or unknown | Seed |
| Torradovirus | Narrow | Whitefly or aphid | unknown |
| Waikavirus | Narrow | Aphid or leafhopper | None |
Antigenicity
Virus preparations are usually good immunogens and polyclonal antibodies prepared against purified virus particles recognize all CPs. Viruses of species belonging to the same genus can be serologically distantly related.
Subfamily demarcation criteria
There is currently only one subfamily (Comovirinae) which groups together three genera (Comovirus, Fabavirus and Nepovirus). These three genera are closely related to each other in phylogenetic analyses (Figure 4 Secoviridae). Given the presence of only one subfamily, formal demarcation criteria have not been defined.
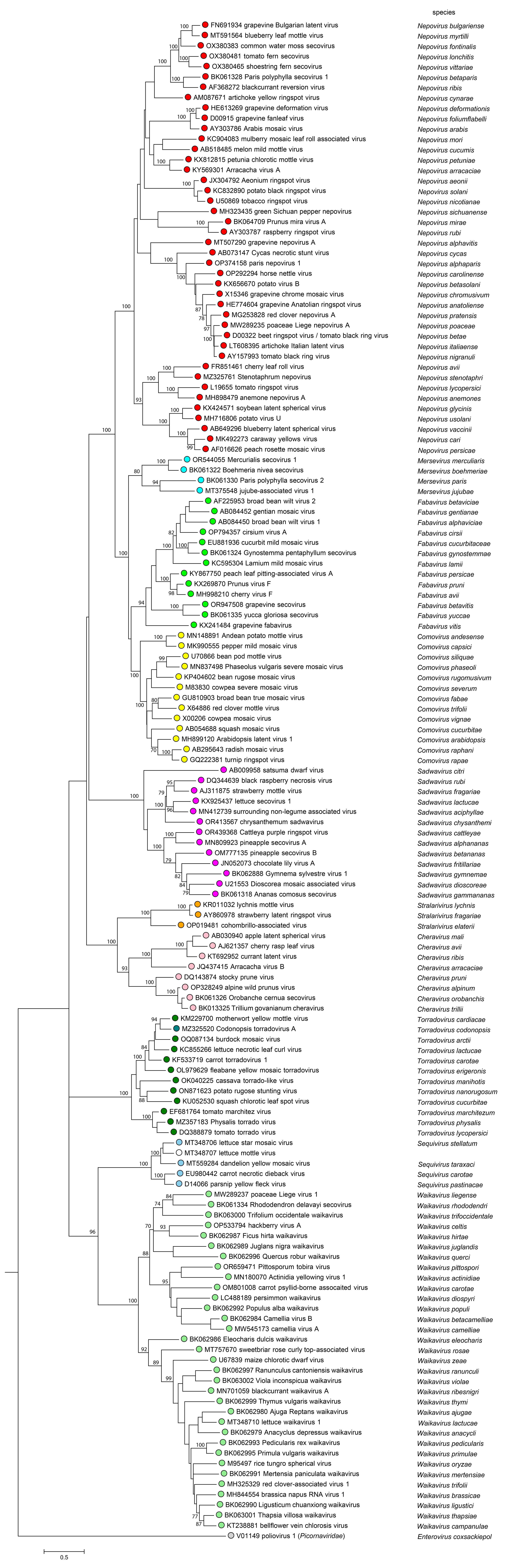 |
| Figure 4 Secoviridae. Phylogenetic tree of members of the family Secoviridae based on an alignment of amino acid sequences of the conserved domains between the “CG” motif of the 3C-proteinase and the “GDD” motif of the polymerase (Pro-Pol region) inferred by using the Maximum Likelihood method and the Le and Gascuel (2008) model. A discrete Gamma distribution was used to model evolutionary rate differences among sites [2 categories (+G, parameter = 1.7191)] and evolutionary analyses were conducted in MEGA X. (Kumar et al., 2018). The bar represents the genetic distance. Bootstrap values > 70% are shown. |
Genus demarcation criteria
The criteria demarcating genera in the family are:
· Number of genomic RNAs
· Number of protein domains and/or processing sites within the polyprotein(s)
· Number of CPs
· Presence of additional ORFs and/or subgenomic RNAs
· Clustering as a single branch in phylogenetic trees derived from amino acid sequence alignments of the conserved Pro-Pol region when compared with other genera of the family Secoviridae (Figure 4 Secoviridae). The Pro-Pol region is delineated by the “CG” motif of the 3C-like proteinase and the “GDD” motif of the polymerase. Identification of proteinase cleavage sites is not required to delineate the Pro-Pol region.
Not all criteria may need to be met simultaneously.
Species demarcation criteria
Useful criteria to demarcate species are:
· CP aa sequence with less than 75% identity (for viruses with two or three CPs, combined CP sequences are considered)
· Conserved Pro-Pol region aa sequence (as defined above) with less than 80% identity
· Differences in antigenic reactions
· Distinct host range
· Distinct vector specificity
· Absence of cross-protection
· For viruses with a bipartite genome, absence of re-assortment between RNA1 and RNA2
Not all criteria need to be met simultaneously. In some cases, sequence information alone can be a good indicator of a distinct species (i.e., when the percentage of aa sequence identity in both the Pro-Pol and CP(s) regions is well below the proposed cut-off). However, analysing only one region of the genome is generally not sufficient and both the Pro-Pol and CP(s) regions should be considered. In cases where the percentage of sequence identity in one or both sequences is near the proposed cut-off (e.g., between 75 and 85% in the Pro-Pol region or between 70 and 80% in the CP(s) region), other criteria should be considered and information on biological properties of the virus (host range, vector specificity, possibility of reassortment between RNAs) is useful. For example, beet ringspot virus (BRSV) and tomato black ring virus (TBRV) (genus Nepovirus) are closely related in the Pro-Pol sequence (89% sequence identity) but are much more divergent in the CP sequence (62% sequence identity). They differ in their antigenic reactions, and also in the specificity of nematode transmission (BRSV is transmitted more efficiently by Longidorus elongatus and TBRV is transmitted more efficiently by Longidorus attenuatus).
Derivation of names
Cheravirus: from cherry rasp leaf virus, a virus in the genus Cheravirus
Comovirinae, Comovirus: from cowpea mosaic virus, a virus in the genus Comovirus
Fabavirus: derived from the Latin faba, bean; also Vicia faba, broad bean
Mersevirus: from Mercurialis secovirus 1, a virus in the genus
Nepovirus: from nematode-transmitted, polyhedral particles
Sadwavirus: from satsuma dwarf virus, a virus in the genus Sadwavirus
Secoviridae: derived from the amalgamation of the previous families Sequiviridae and Comoviridae.
Sequivirus: from Latin sequi, to follow, accompany (in reference to the dependent aphid transmission of parsnip yellow fleck virus)
Stralarivirus: from strawberry latent ringspot virus, a virus in the genus Stralarivirus
Torradovirus: derived from tomato torrado virus, a virus in the genus Torradovirus. In Spanish, torrado means toasted to refer to the severe necrosis (burnt-like phenotype) observed in the disease induced by tomato torrado virus.
Waikavirus: from Japanese, describing the symptoms induced in rice by infection with rice tungro spherical virus alone (i.e. in the absence of rice tungro bacilliform virus)
Relationships within the family
Members of the family Secoviridae were previously classified in two different families: Comoviridae (including the genera Comovirus, Fabavirus and Nepovirus) and Sequiviridae (including the genera Sequivirus and Waikavirus) and in two unassigned genera: Cheravirus and Sadwavirus. The families and genera were amalgamated to create the new family Secoviridae, which includes all plant viruses that are members of the order Picornavirales (Sanfaçon et al., 2009).
The conserved Pro-Pol region on RNA1, delineated by the “CG” motif of the 3C-like proteinase and the “GDD” motif of the polymerase, has been used to determine the relationship among members of the order Picornavirales. Comparison of the Pro-Pol sequence among members of the family Secoviridae allows the definition of branches that generally correspond to the distinct genera. Members of the sub-family Comovirinae (genera Comovirus, Fabavirus and Nepovirus) are more closely related to each other than to other genera within the family (Figure 4.Secoviridae). Within this sub-family, fabaviruses and comoviruses are more closely related to each other than to nepoviruses.
Relationships with other taxa
Members of the family Secoviridae are related to members of other families in the order Picornavirales (Thompson et al., 2014, Thompson 2020). They all share a common virion structure, organization of the replication block within the polyproteins and conserved properties of the replication proteins, including the 3C-like proteinase. Members of the family Secoviridae are also related to members of the families Potyviridae and Caliciviridae in some aspects (common replication block, polyprotein expression strategy, VPg bound to the 5ʹ-end of the genome, and poly(A) tail at the 3ʹ-end of the genome) but differ in other properties.