Family: Tristromaviridae
David Prangishvili, Elena Rensen, Tomohiro Mochizuki and Mart Krupovic
The citation for this ICTV Report chapter is the summary published as Prangishvili et al., (2018):
ICTV Virus Taxonomy Profile: Tristromaviridae, Journal of General Virology, 100, 135–136.
Corresponding authors: David Prangishvili ([email protected]) and Mart Krupovic ([email protected])
Edited by: Andrew M. Kropinski and Stuart G. Siddell
Posted: October 2018
PDF: ICTV_Tristromaviridae.pdf
Summary
Tristromaviridae is a family of viruses with linear, double-stranded DNA genomes of 16–18 kbp (Table 1. Tristromaviridae). The flexible, filamentous virions (400 ± 20 nm × 30 ± 3 nm) consist of a lipid envelope and an inner core constructed from two structural units: a rod-shaped helical nucleocapsid and a nucleocapsid-encompassing matrix protein layer. Tristromaviruses are lytic and infect hyperthermophilic archaea of the order Thermoproteales.
Table 1. Tristromaviridae. Characteristics of members of the family Tristromaviridae.
| Characteristic | Description |
| Typical member: | Pyrobaculum filamentous virus 1 (KU307456), species Alphatristromavirus pozzuoliense |
| Virion | Filamentous, flexible (400 ± 20 nm × 30 ± 3 nm) particle consisting of a nucleoprotein core covered by a matrix protein layer and enveloped with a lipid membrane; bundles of thin filaments are attached to both ends |
| Genome | Linear, double-stranded DNA genomes of 16–18 kbp |
| Replication | Viruses are lytic; virions are released by the rupture of the host cell envelope |
| Translation | Not characterized |
| Host range | Hyperthermophilic archaea of the order Thermoproteales |
| Taxonomy | Realm Adnaviria, kingdom Zilligvirae, phylum Taleaviricota, class Tokiviricetes, order Primavirales: two genera, three species |
Virion
Morphology
The virions are filamentous, 400 ± 20 × 32 ± 3 nm, and contain a lipid-containing envelope and an inner core consisting of two structural units: (i) a rod-shaped helical nucleocapsid, formed of two major virion proteins (VP1 and VP2), and (ii) a nucleocapsid-encompassing protein (VP3) sheath (Table 1. Tristromaviridae, Figure 1. Tristromaviridae) (Rensen et al., 2016). The sheath layer is sandwiched between the nucleocapsid and the envelope, akin to the matrix protein layer found in certain eukaryotic negative-sense RNA viruses, e.g. members of the Filoviridae (King et al., 2012). Filamentous viruses with dsDNA genomes are thus far exclusive to hosts in the domain Archaea (Prangishvili et al., 2017).
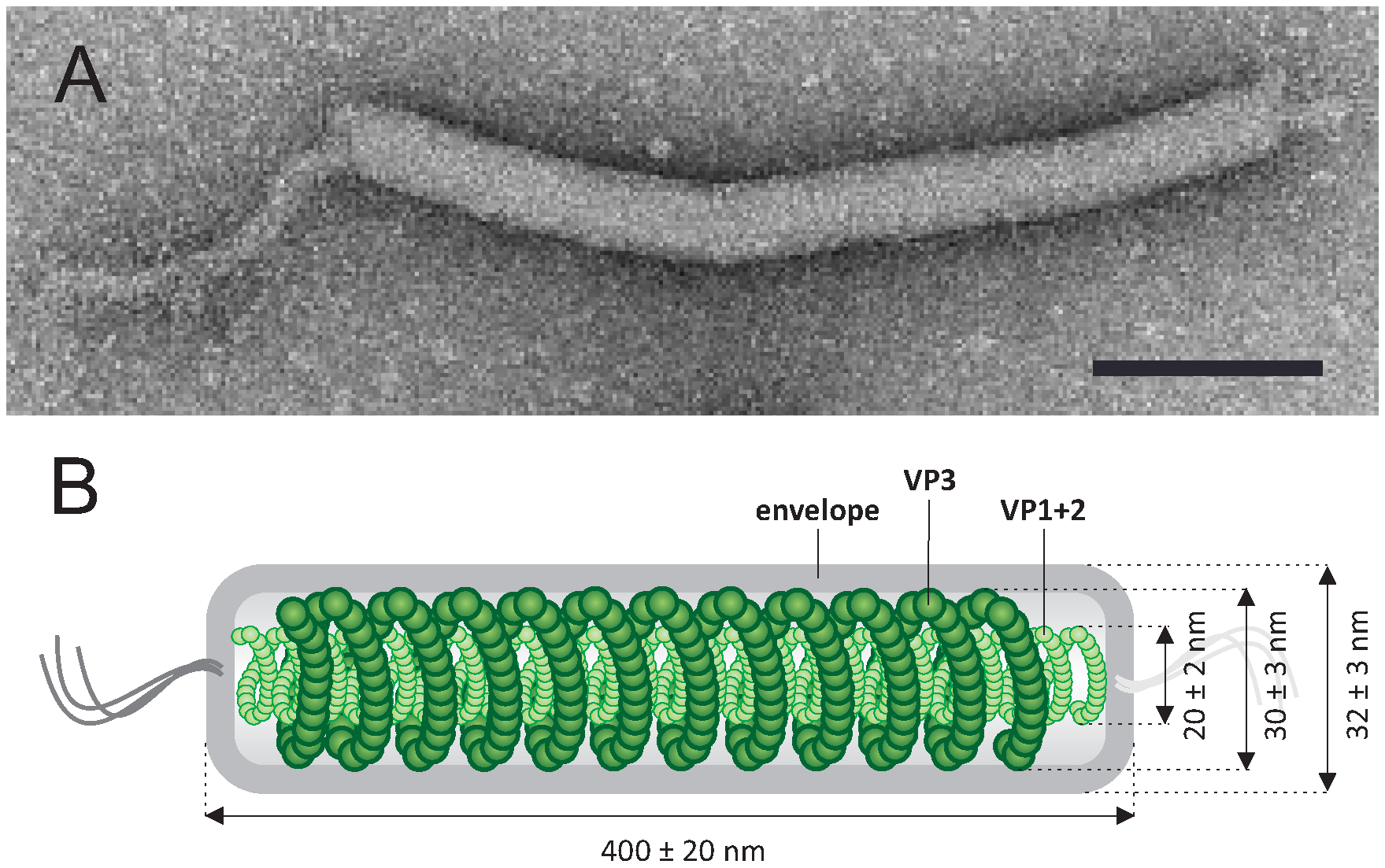 |
| Figure 1. .Tristromaviridae. Tristromaviridae. (A) Electron micrograph of a virion of negatively-stained Pyrobaculum filamentous virus 1, stained with 2% uranyl acetate. (B) A schematic of the virion organization with the position of 3 major capsid proteins indicated. Scale bar, 100 nm. Modified from (Rensen et al., 2016). |
Physicochemical and physical properties
Non-ionic detergents, such as Triton X-100 and Nonidet P-40, dissociate virions into viral cores, containing the dsDNA genome and the DNA-binding nucleocapsid proteins, and the viral envelopes containing lipids. Mild treatment with Tween-20 causes partial detachment of the envelope from the nucleocapsid but not its complete removal. The virions are completely degraded when treated with 0.1% SDS. Treatment with ethanol (50% vol/vol) does not affect the integrity of the virions but results in their conversion into cane-shaped particles. Virions appear to be sensitive to mechanical stress and can be partially disrupted in the course of purification and concentration (Rensen et al., 2016).
Nucleic acid
The linear dsDNA genome of Pyrobaculum filamentous virus 1 (PFV1) is 17,714 bp, includes 60 bp terminal inverted repeats, and is predicted to encode 39 proteins (Figure 2. Tristromaviridae), most of which do not show similarities to sequences in public databases (Krupovic et al., 2018). The genome has a G+C content of 45.4%, which is considerably lower than that of the host genome (~55%).
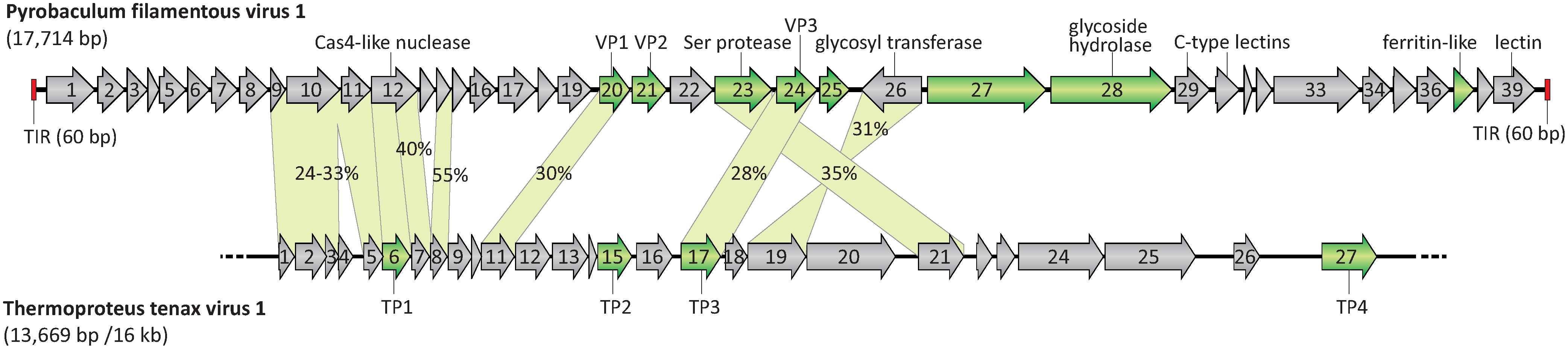 |
| Figure 2. Tristromaviridae. A genome map of Pyrobaculum flamentous virus 1 compared with the partial genome map of the virus Thermoproteus tenax virus 1. ORFs are shown by arrows, which also indicate the direction of transcription. Genes encoding the structural proteins are shown in green. Terminal inverted repeats (TIR) are depicted by red rectangles. The incompleteness of the Thermoproteus tenax virus 1 genome is illustrated by the broken line. TP: Thermoproteus tenax virus 1 capsid protein. The genes shared by both viruses are connected by green shading, and the identity between the corresponding proteins is indicated. Modified from (Rensen et al., 2016). |
Proteins
PFV1 virions contain three major capsid proteins, VP1, VP2, and VP3, with molecular masses of 14 kDa, 14 kDa, and 18 kDa, respectively, and at least five minor proteins with molecular masses in the range of 11–30 kDa (Figure 2. .Tristromaviridae). The proteins VP1 and VP2 bind the viral dsDNA and form the nucleoprotein filament. The highly hydrophobic 18 kDa protein, VP3, forms a nucleocapsid-encompassing protein sheath and is glycosylated (Figure 1. Tristromaviridae) (Rensen et al., 2016). The virions of the only other known tristromavirus, Thermoproteus tenax virus 1 (TTV1; Figure 3. Tristromaviridae), also include three major structural proteins, but only one of these, namely TP3, is shared with PFV1. Although some of the minor structural proteins of PFV1 have homologs in the TTV1 genome (Figure 2. Tristromaviridae), they have not been identified in TTV1 virions (Janekovic et al., 1983). Notably, one of the two major nucleocapsid proteins of TTV1, protein TP1, has evolved from a truncated Cas4-like nuclease (Krupovic et al., 2015). The gene for the Cas4-like nuclease is intact in the PFV1 genome (Figure 2. Tristromaviridae).
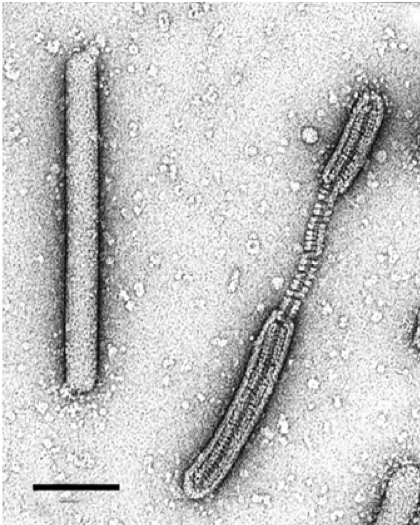 |
| Figure 3. Tristromaviridae. Electron micrograph of intact and partially deteriorated virions of negatively-stained Thermoproteus tenax virus 1, stained with 2% uranyl acetate. Scale bar, 100 nm. |
Lipids
The envelope of the virion contains lipids derived from the pool of host lipids: glycerol dibiphytanyl glycerol tetraether lipids and small amounts of the glycerol trialkyl glycerol tetraether lipids (Rensen et al., 2016).
Carbohydrates
The sheath protein VP3 of PFV1 has been shown to be glycosylated but the exact structure of the glycan remains unknown (Rensen et al., 2016).
Genome organization and replication
The PFV1 genome is predicted to encode 39 proteins that are larger than 40 amino acids (Figure 2. Tristromaviridae) (Rensen et al., 2016). The genes are tightly arranged and occupy 93.1% of the genome. All except one of the genes are encoded on the same strand. Thirty-six genes use AUG as a start codon. Ten of the PFV1 genes were predicted to encode proteins with one or more transmembrane domains. Sequence analyses allowed the assignment of putative functions to less than a quarter of all PFV1 genes (Krupovic et al., 2018). Five of them were predicted to be involved in carbohydrate metabolism, one encodes a Cas4-like nuclease, and others represent a ferritin protein and a putative trypsin-like serine protease (Figure 2. Tristromaviridae).
Nine gene products of PFV1 share significant sequence similarity with proteins encoded in the partially sequenced genome of the virus TTV1 (Neumann et al., 1989) (Figure 2. Tristromaviridae). Two of these genes are split in TTV1 when compared to PFV1.
Biology
The virus PFV1 was isolated from its host, obtained from a hot spring (87–93 °C, pH 6) in Pozzuoli, Italy (Rensen et al., 2016). The host range is limited to members of the hyperthermophilic archaea from the genus Pyrobaculum (order Thermoproteales). The virus TTV1 infects Thermoproteus tenax (genus Thermoproteus, order Thermoproteales) and was isolated with its host from a mud hole of 93°C, pH 6 at Krafla, Iceland (Janekovic et al., 1983). PFV1 binds to the host cell via interaction of the terminal protrusions of the virion with the host pili-like appendages. The infection leads to cell lysis. Mature virions appear to assemble in the host cell lumen prior to release, which coincides with the rupture of the host cell envelope. The virus does not encode identifiable DNA and RNA polymerases and is likely to depend on the host machineries for genome replication and transcription.
Derivation of names
Tristroma: from Greek tria for three and stroma for a layer
Relationships within the family
TTV1 was originally assigned to the family Lipothrixviridae (genus Alphalipothrixvirus) based on its overall morphology and the presence of the external envelope. However, since subsequent comparative genomics analysis has shown that TTV1 does not share any homologous genes with other members of the Lipothrixviridae, the virus has been reclassified into the family Tristromaviridae (Rensen et al., 2016).

