Family: Belpaoviridae
Beatriz Soriano, Mart Krupovic and Carlos Llorens
Summary
Belpaoviridae is a family of reverse-transcribing viruses with long terminal repeats (LTRs), commonly known as Bel/Pao LTR retrotransposons, and found in metazoans (Table 1.Belpaoviridae). The family Belpaoviridae belongs to the order Ortervirales together with other families of reverse-transcribing viruses, namely Metaviridae, Pseudoviridae, Retroviridae and Caulimoviridae, with which they share evolutionary history and functional and structural features. Belpaoviruses were originally classified within the genus Semotivirus in the family Metaviridae. However, based on phylogenetic evidence that metavirids and belpaovirids segregate into two monophyletic groups, the genus Semotivirus has been moved to become the only genus in the family Belpaoviridae. A number of unclassified members of the family may represent additional genera.
Table 1. Belpaoviridae. Characteristics of members of the family Belpaoviridae
| Characteristic | Description |
| Example | Ascaris lumbricoides Tas virus (Z29712), species Semotivirus tasascaridis |
| Virion | Unknown |
| Genome | Positive-sense linear ssRNA of 4–10 kb |
| Replication | By reverse-transcription primed by a host-encoded tRNA |
| Translation | Genomic RNA is translated into one or more polyproteins |
| Host range | Vertebrates, insects and nematodes |
| Taxonomy | Realm Riboviria, kingdom Pararnavirae, phylum Artverviricota, class Revtraviricetes, order Ortervirales; the genus Semotivirus includes 11 species |
Virion
Morphology
Little is known about the virion morphology of belpaovirids. However, given that belpaovirids encode a Gag polyprotein with nucleocapsid and capsid protein domains homologous to those of other members of the order Ortervirales (Krupovic and Koonin 2017, Krupovic et al., 2018), their replication probably involves the formation of virus-like particles (VLPs), as in the case of retrovirids, metavirids and pseudovirids. Some members of the family Belpaoviridae, such as Ascaris lumbricoides Tas virus (AluTasV) and Drosophila melanogaster Roo virus (DmeRooV), have an env-like gene (de la Chaux and Wagner 2011), the function of which remains unknown.
Physicochemical and physical properties
No information is available.
Nucleic acid
By analogy with members of the families Metaviridae and Pseudoviridae, it is assumed that the VLPs of belpaovirids package two copies of a positive-sense ssRNA genome as well as some cellular tRNAs. Genome lengths range from 4.2 to 10 kb.
Proteins
Belpaovirids encode Gag and Pol polyproteins, which is typical of all LTR retrotransposons. Gag is usually processed into the major structural proteins: the capsid protein (CP) and the nucleocapsid (NC) protein. By functional analogy with retroviruses, the CP of belpaovirids is thought to form the immature VLP, while the NC packages the RNA genome within the VLP. In terms of protein domain architecture, the C-terminus of the NCs of almost all Belpaoviridae species present three copies of an Cys-X2-Cys-X4-His-X4-Cys (CCHC) zinc knuckle motif similar to that observed in the NCs of other viruses in the order Ortervirales (Llorens et al., 2009b).
The Pol polyprotein is processed into the characteristic protease (PR), reverse transcriptase-ribonuclease H (RT-RH) and integrase (INT) enzymes necessary for the RNA-to-cDNA reverse transcription and integration steps. At the protein architecture level, PR has the typical DTG/ILG consensus sequence domain of clan AA proteases (Llorens et al., 2009a); the RT-RH domains include the conserved regions common to RTs of all LTR retroelements; and INT displays the canonical His-X5-His-Xn-Cys-X3-Cys zinc finger motif followed by the DD35E-like core, which is also common to all retroviral DDE INTs (Llorens et al., 2009b). While most belpaovirids are LTR retrotransposons, some, such as Ascaris lumbricoides Tas virus (AluTasV) and Drosophila melanogaster Roo virus (DmeRooV), carry a third ORF with the typical domain features of retroviral envelope (Env)-like proteins. These belpaovirids may form infectious extracellular virions, akin to some metavirids, although there is currently no experimental evidence for this.
Lipids
No information is available.
Carbohydrates
No information is available.
Genome organization and replication
Members of the family Belpaoviridae normally have a genomic organization typical of LTR retrotransposons, with 1−3 genes (gag, pol and env) being flanked by LTRs (Llorens et al., 2011). The length of the full-length genome varies from 4.2 to 10 kb. The length of the LTRs is also variable and, depending on the species, ranges from 0.2 to 1.2 kb. Downstream of the 5′-LTR, there is a non-coding region of variable length that corresponds to the first portion of the reverse-transcribed genome. A primer-binding site (PBS) of 18 nt is localized downstream from this non-coding region. The PBS is complementary to a specific region within the 3′-end of a host tRNAArg or tRNAGly. Upstream of the 3′-LTR is a small region of approximately 10 A/G residues called the polypurine tract (PPT), which is responsible for starting the synthesis of the proviral DNA strand. The Gag and Pol polyproteins can be encoded by one continuous or two overlapping gag and pol genes (Figure 1. Belpaoviridae). The gag gene is located upstream of the pol gene and encodes the typical CP and NC domains. However, in some viruses, such as Drosophila melanogaster Max virus (DmeMaxV), a partial duplication of gag is found downstream of the pol gene. The pol gene is located downstream of the gag gene, and encodes the Pol protein with PR, RT-RH and INT domains. In some genomes, the INT domain is expressed from an overlapping reading frame (Marsano and Caizzi 2005). Similar to members of the families Metaviridae and Pseudoviridae, it is believed that PR of belpaovirids is needed to process the Gag polyprotein, whereas RT-RH is involved in reverse transcription synthesis of cDNA from the ssRNA template and the hydrolysis of the original RNA template that is part of the RNA/DNA hybrid generated during reverse transcription. INT catalyzes the insertion of the cDNA copy of the belpaoviral genome into the host chromosome.
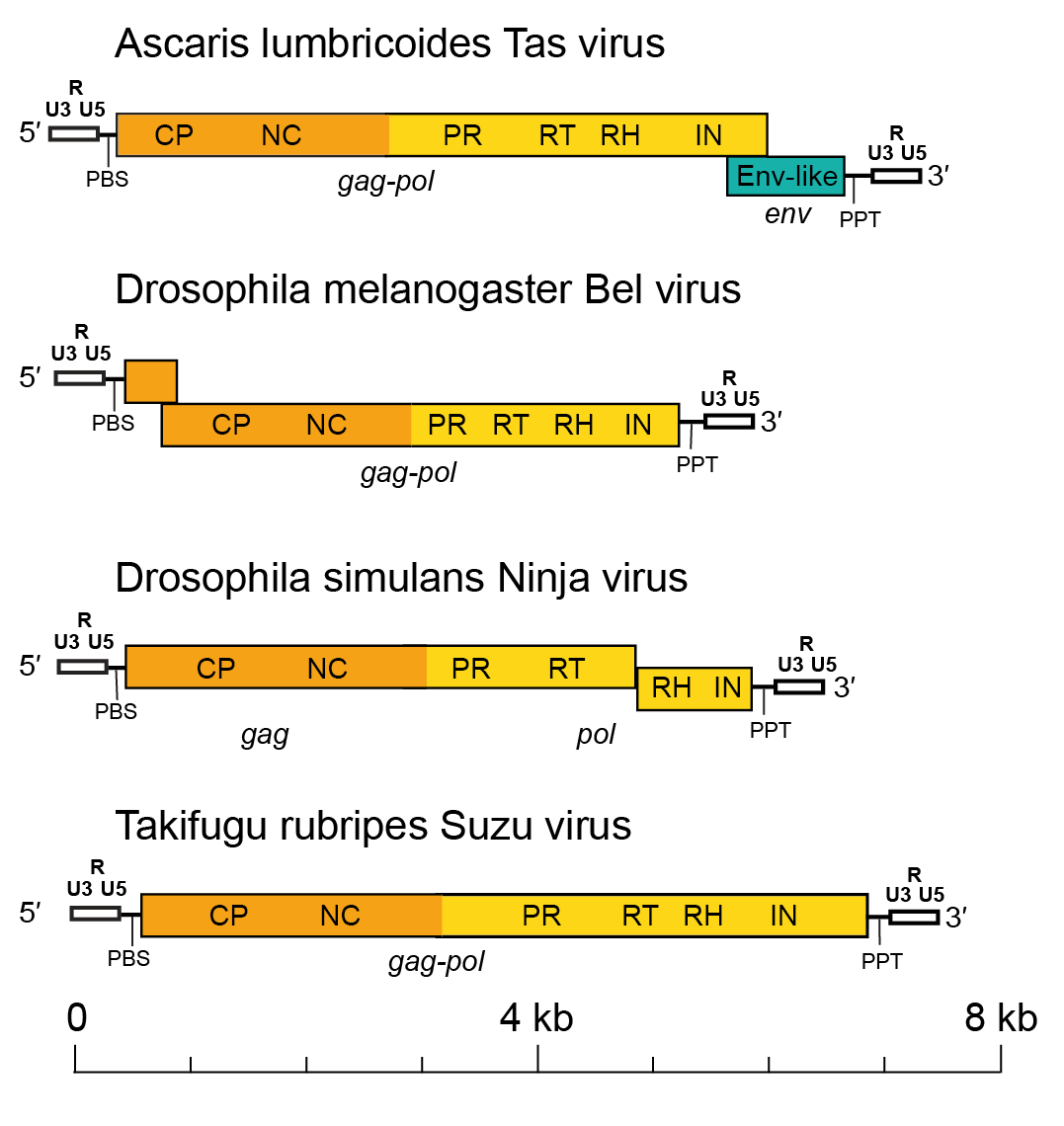 |
| Figure 1. Belpaoviridae. Full-length genome architectures of representative semotiviruses. LTRs, including the U3, R and U5 regions, and the distinct gag, pol, and env regions are differentially colored. The LTRs are white, gag is orange, pol is yellow and env is green. Abbreviations: LTR, long terminal repeats; PBS, primer-binding site; PPT, polypurine tract; CP, capsid protein domain; NC, nucleocapsid protein domain; PR, protease; RT, reverse transcriptase; RH, ribonuclease H; INT, integrase; and Env-like, envelope-like protein. |
In comparison to other families of the order of Ortervirales, the order of Pol domains PR-RT-RH-INT in belpaovirids is the same as that observed in Pol polyproteins of members of the families Metaviridae and Retroviridae. This Pol domain organization differs from that observed in members of the family Pseudoviridae, in which INT is located upstream of PR, or in members of the family Caulimoviridae, which lack the INT domain.
The genome replication mechanism of members of the family Belpaoviridae is poorly understood, but, given the similarity in both genome structure and protein domains, it is assumed to be similar to that of members of the family Metaviridae (see (Llorens et al., 2020) and the ICTV Report on the family Metaviridae for more details). That is, RT mediates the conversion of full transcripts into dsDNA, which are integrated into the host genome by the INT protein. The host RNA polymerase II then transcribes integrated provirus to form new virus mRNAs, which are capped and polyadenylated by host enzymes, exported to the cytoplasm and translated to produce the Gag and Pol polyproteins that form immature VLPs. The polyproteins are subsequently proteolytically processed by the viral PR, resulting in VLP maturation. RT reverse-transcribes the new viral RNAs to produce dsDNA molecules, which are transported back to the nucleus where they can be inserted at new sites in the host cell genome (Boeke 2013).
Biology
Members of the family Belpaoviridae occur in metazoan genomes, in which they are widely distributed. A study focusing on Bactrocera oleae Achilles virus (BaAchV), an unclassified virus in the family, revealed that its genome is distributed in discrete regions dispersed on all five host autosomes, in all centromeric regions and in the granular heterochromatic network corresponding to the mitotic sex chromosomes, particularly on the Y chromosome (Tsoumani et al., 2015). Another study focusing on the dynamics of members of the family Belpaoviridae, specifically focused on bel-like and ninja-like virus within the Aedes aegypti genome, also showed that provirus was predominantly distributed in intergenic regions and within introns (Minervini et al., 2009). Interestingly, belpaovirids are particularly abundant in nematodes; the Caenorhabditis elegans genome has 19 distinct LTR retroelement lineages, of which 13 are belpaovirids (Ganko et al., 2001), although the majority of these are no longer active. An unusual feature of many of the C. elegans belpaovirid sequences is the presence of additional DNA between the 5′-LTR and the beginning of the first ORF. These additional sequences are variable within the same group of elements and completely different between groups. One active group of sequences, represented by Caenorhabditis elegans Cer13 virus (CelCer13V), also contains a third env-like gene located between the pol gene and the 3′-LTR. This putative Env-like protein exhibits no sequence similarity to the corresponding protein in AluTasV, suggesting an independent acquisition. The likely origins of the AluTasV and CelCer13V Env-like proteins are the gB and G2 glycoproteins of herpesviruses and phleboviruses, respectively.
Derivation of names
Belpaoviridae: from the names of the LTR retrotransposon clades Bel and Pao, in turn derived from the virus names Drosophila melanogaster Bel virus and Bombyx mori Pao virus, two representative members of the family
Semotivirus: from Latin semotus, meaning distant, removed. This prefix refers to the observation that, based on the sequence of the RT domain, the viruses in this genus are distantly related to members of the family Metaviridae, the family in which the genus was initially placed.
Genus demarcation criteria
There is only one genus in the family.
Species demarcation criteria
Members of different species are less than 50% identical in their Gag protein sequence.
Relationships within the family
Phylogenetic analysis of the most conserved part of the RT domain in classified and potential members of the family Belpaoviridae reveals the presence of five clades, here designated as Tas, Bel, Pao, Sinbad and Suzu (Figure 2. Belpaoviridae). These clades correlate with the host of origin as follows: Pao – invertebrates and vertebrates, Suzu – marine vertebrates and echinoderms, Tas – cnidarians and nematodes, Sinbad – protostome and deuterostome worms, and Bel – insects. Similar phylogenies are observed from analysis of the RH and INT domains, and also for the less well conserved Gag and PR proteins (Llorens et al., 2009b, Llorens et al., 2011, Llorens et al., 2008); further information is available at the Gypsy Database (GyDB) (http://gydb.org).
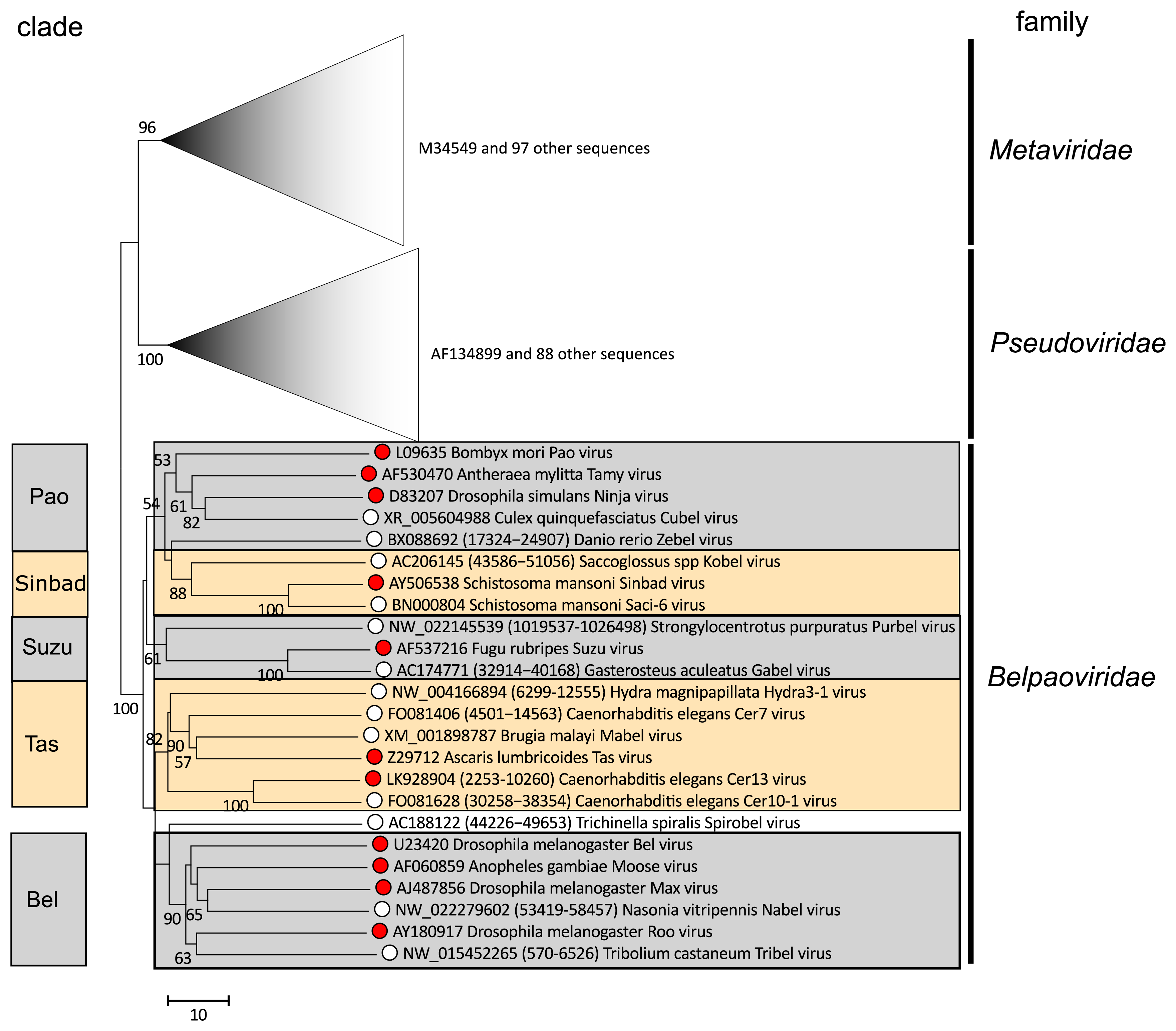 |
| Figure 2. Belpaoviridae. Phylogenetic tree of the Belpaoviridae family based on the alignment of the RT core of 210 operational taxonomic units (OTUs) belonging to the Metaviridae, Pseudoviridae and Belpaoviridae families. ClustalW (Larkin et al., 2007) and GeneDoc (https://genedoc.software.informer.com/2.7) were used to align and manually refine the sequences. Clustal W was also used to perform the phylogenetic reconstruction analysis using the neighbor-joining method and 1000 bootstrap replicates (values >50% shown). The branches corresponding to the Metaviridae and Pseudoviridae families are collapsed and used to root the tree. Red spots indicate members of Belpaoviridae species; open circles indicate potential members, corresponding to the set of non-redundant canonical belpaovirids retrieved from the GyDB, are summarized in the section Related, unclassified viruses. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Members of the family Belpaoviridae are most closely reated to members of other families in the order Ortervirales, namely the families Metaviridae, Pseudoviridae, Retroviridae and Caulimoviridae. Because of the similarity to Metaviridae family in both sequence and functional features, it is commonly assumed that the family Belpaoviridae evolved from a metavirus ancestor probably after the split of fungi and metazoans (Llorens et al., 2009b).
Related, unclassified viruses
| Clade | Virus name | Accession number |
| Bel | Tribolium castaneum Tribel virus | NW_015452265 (570–6526) |
| Bel | Nasonia vitripennis Nabel virus | NW_022279602 (53419–58457) |
| Tas | Caenorhabditis elegans Cer10-1 virus | FO081628& (30258–38354) |
| Tas | Trichinella spiralis Spirobel virus | AC188122 (44226–49653) |
| Tas | Hydra magnipapillata Hydra3-1 virus | NW_004166894 (6299–12555) |
| Tas | Caenorhabditis elegans Cer7 virus | FO081406 (4501–14563) |
| Tas | Brugia malayi Mabel virus | XM_001898787 |
| Suzu | Strongylocentrotus purpuratus Purbel virus | NW_022145539 (1019537–1026498) |
| Suzu | Gasterosteus aculeatus Gabel virus | AC174771 (32914–40168) |
| Sinbad | Saccoglossus spp Kobel virus | AC206145 (43586–51056) |
| Sinbad | Schistosoma mansoni Saci-6 virus | BN000804 |
| Pao | Danio rerio Zebel virus | BX088692 (17324–24907) |
| Pao | Culex quinquefasciatus Cubel virus | XR_005604988 |
| Bactrocera oleae Achilles virus | KT280063 |
Numbers in parentheses are positions of virus sequences within a larger sequence

