Family: Fusariviridae
Sotaro Chiba, Nobuhiro Suzuki, Leonardo Velasco, María A. Ayllón, Shin-Yi Lee-Marzano, Liying Sun, Sead Sabanadzovic, and Massimo Turina
The citation for this ICTV Report chapter is the summary published as:
Corresponding author: Massimo Turina (massimo.turina@ipsp.cnr.it)
Edited by: Sead Sabanadzovic and Evelien Adriaenssens
Posted: March 2024
Summary
Fusariviridae is a family of mono-segmented, positive-sense RNA viruses with a genome size of 5.9–10.7 kb (Table 1 Fusarividae). The family Fusarividae includes 34 species, grouped in the three genera Alphafusarivirus, Betafusarivirus and Gammafusarivirus. In the vast majority of cases, genomic RNAs are bicistronic, with some exceptions where up to four predicted ORFs are present. In the bicistronic fusarivirids, the 5′-proximal ORF codes for a single protein with both RNA-directed RNA polymerase (RdRP) and RNA helicase (Hel) domains; little is known about the protein encoded by the second ORF. The 3′-terminus of the genomic RNA is generally polyadenylated. Fusarivirids do not appear to form virions; during their replication cycle they accumulate intracellularly an abundance of the replicative form of dsRNA.
Table 1 Fusariviridae. Characteristics of members of the family Fusariviridae
| Characteristic | Description |
| Example | Fusarium graminearum dsRNA mycovirus 1 (AY533037), species Alphafusarivirus fusarii, genus Alphafusarivirus |
| Virion | Thought to be capsidless since no virions can be purified on a sucrose density gradient |
| Genome | 5.9–10.7 kb mono-segmented positive-sense RNA |
| Replication | Unknown |
| Translation | Expression of Fusarium graminearum dsRNA mycovirus-1 downstream ORFs is from subgenomic RNAs. No evidence for such molecules in Rosellinia necatrix fusarivirus 1 |
| Host range | Experimentally confirmed hosts are members of the kingdom Fungi and in one case a confirmed stramenopiles oomycetes host. Additional members of two species have been associated with peronosporaceaen stramenopiles from a metatranscriptomic sample. |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Pisuviricota, class Duploviricetes, order Durnavirales: 3 genera and 34 species |
Virion
Morphology
No true virions are associated with fusarivirids. No infectious form has been identified yet.
Nucleic acid
The members of this family characterized so far have positive-sense RNA genomes of 5.9 kb to 10.7 kb
Genome organization and replication
Viruses belonging to the family have very diverse genome organizations varying from monocistronic (Lentinula edodes fusarivirus 1) (Liu et al., 2023) or bicistronic (Rosellinia necatrix fusarivirus 1) (Zhang et al., 2014) to more complex genome organizations with 3 (Pleospora tiphycola fusarivirus 1) (Nerva et al., 2016) or 4 ORFs (Rhizoctonia solani fusarivirus 1) (Picarelli et al., 2019) (Figure 1 Fusariviridae). In all genomes, the largest ORF codes for a single protein with both RNA-directed RNA polymerase (RdRP) and RNA helicase (Hel) domains. The second ORF encodes a conserved protein with unknown function, although it contains a motif from a structural maintenance of chromosomes (SMC) protein (PDB entry code: 6YUF) (Gao et al., 2021, Sahin et al., 2021). Accessory ORFs are often genus-specific or have no detectable similarity with those of any other viruses. In the largest quadricistronic fusarivirids a second helicase domain is present in the protein encoded by ORF1, and this helicase is more similar to those of potyvirids and hypovirids, suggesting possible horizontal gene transfer between these viruses (Abdoulaye et al., 2022).
Expression of ORFs2–4 of Fusarium graminearum dsRNA mycovirus 1 (FgV1) is likely to occur through the production of subgenomic RNAs (Kwon et al., 2007).
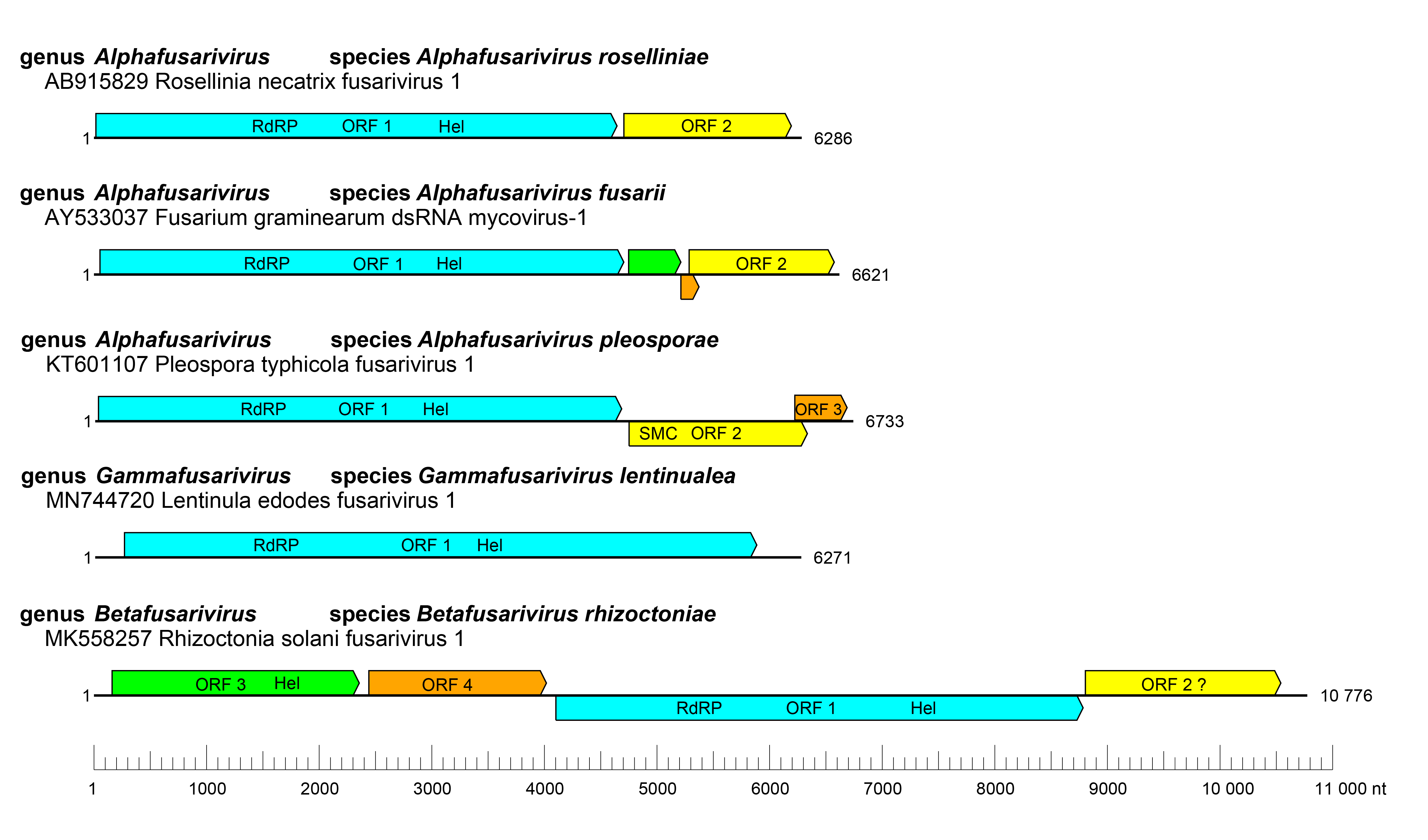 |
| Figure 1 Fusariviridae. Genome organisation of representative fusariviruses. RdRP, RNA-directed RNA polymerase domain, Hel, Helicase domain, SMC, structural maintenance of chromosomes domain. |
Biology
Fusarivirids infect ascomycetes and basidiomycetes in the kingdom Fungi. Pythium ultimum RNA virus 1 infects the stramenopiles oomycetes Globisporangium ultimum (Fukunishi et al., 2021).
A Fusarium graminearum gene product, hexagonal peroxisome (Hex1) protein, was identified as necessary for efficient FgV1 replication and symptom induction (Son et al., 2013). Hex1 is a major component of the Woronin body, a peroxisome-derived organelle that prevents the extension of wound-induced damage to neighboring cells. The involvement of Hex1 was also investigated on Neurospora crassa fusarivirus 1 (NcFV1) replication in N. crassa, and no discernible effect was observed, suggesting that the involvement of Hex1 in fusarivirus replication is specific to the pathosystem F. graminearum-FgV1 (Honda et al., 2020).
A comparison of colony morphology between virus-free and virus-infected isogenic strains grown on nutrition-rich (PDA) and -poor (Vogel's) media suggests that infections of Rosellinia necatrix with RnFV1 is asymptomatic. Results were confirmed with a virulence assay that showed no significant difference between these fungal strains on apple rootstocks (Zhang et al., 2014). Lack of symptoms was also observed by transmission of the virus to two independent virus-free Sclerotinia sclerotiorum strains and one infected with Sclerotinia sclerotiorum fusarivirus 1 (SsFV1) (Liu et al., 2015). In the case of FgV1 strain SD4 (FgV1-SD4), isogenic virus-infected and virus-free strains demonstrated a role for the virus in hypovirulence by eliminating deoxynivalenol synthesis in F. graminearum (Paudel et al., 2022).
Penicillium aurantiogriseum fusari-like virus 1 (PaFV1) has been transfected into plant protoplasts but without evidence of replication in that heterologous system (Nerva et al., 2017).
Derivation of names
Fusariviridae: from the host of the first virus characterized of this family (Fusarium graminearum).
Genus demarcation criteria
Members of different genera belong to monophyletic clades in phylogenetic trees derived from an alignment of the protein that includes the RdRP domain (Figure 2 Fusariviridae).
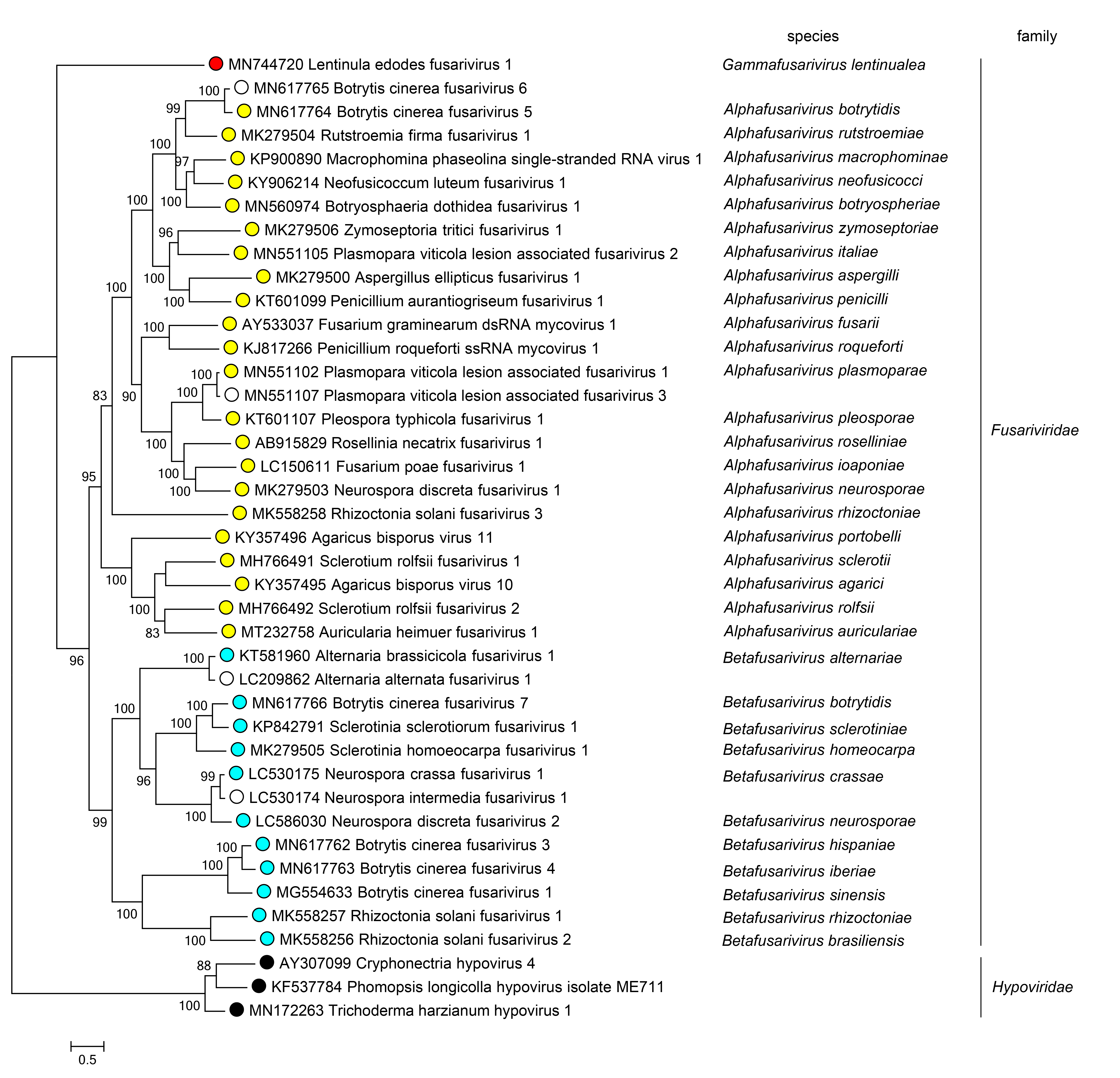 |
| Figure 2 Fusariviridae. Phylogenetic analysis of fusarivirus RdRP protein sequences. Numbers at nodes indicated percentage bootstrap support. Representative sequences of the family Hypoviridae are used as outgroup. |
Species demarcation criteria
Viruses belonging to the same species are >80% identical in the full-length sequence of the protein that includes an RdRP domain.
Relationships within the family
Three major monophyletic groups, corresponding to the three genera in the family, can be identified by phylogenetic analysis of the complete sequence of the protein that includes the RdRP domain (Figure 2 Fusariviridae).
Relationships with other taxa
Deduced amino acid sequences of the RdRP and Hel domains of viruses in the family Fusariviridae place them as a distinct clade close to viruses in the family Hypoviridae (Kwon et al., 2007, Zhang et al., 2014). However, fusarivirids have a different genome organization and relative position of the RdRP and Hel domains. Furthermore, some groups of viruses that putatively code for a coat protein have been detected in metatranscriptomic studies and are basal to members of the family Hypoviridae in phylogenetic analyses. The largest of such groups (identified as f.066) is populated with fusarivirids suggesting that a coat protein could have been present in the ancestors of fusarivirds (Neri et al., 2022).
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Alternaria solani fusarivirus 1 | MW544173 | AsFV1 |
| Botryosphaeria dothidea fusarivirus 2 | MW863557 | BdFV2 |
| Botrytis cinerea fusarivirus 8 | OK558541 | BcFV8 |
| Botrytis cinerea fusarivirus 9 | OK558542 | BcFV9 |
| Corynespora cassiicola fusarivirus 1 | OL456888 | CcFV1 |
| Erysiphe necator associated fusarivirus 1 | MN558695 | EnFV1 |
| Fusarium fusarivirus 1 | OP729394 | FuFV1 |
| Fusarium pseudograminearum fusarivirus 1 | OP100308 | FpsFV1 |
| Gaeumannomyces tritici fusarivirus 1 | MK279501 | GtFV1 |
| Morchella esculenta fusarivirus 1 | MW965472 | MeFV1 |
| Morchella esculenta fusarivirus 2 | MW965473 | MeFV2 |
| Monilinia fructicola fusarivirus 1 | ON038402 | MfFV1 |
| Monilinia fructicola fusarivirus 2 | ON038403 | MfFV2 |
| Morchella importuna fusarivirus 1 | MK279502 | MiFV1 |
| Nigrospora oryzae fusarivirus 1 | KU980909 | NoFV1 |
| Nigrospora sphaerica fusarivirus 1 | MT774523 | NsFV1 |
| Rhizoctonia solani fusarivirus 4 | MZ636366 | RsFV4 |
| Rhizoctonia solani fusarivirus 5 | MW713065 | RsFV5 |
| Rhizoctonia solani fusarivirus 6 | MW149059 | RsFV6 |
| Setosphaeria turcica fusarivirus 1 | MW489564 | StFV1 |
Virus names and virus abbreviations are not official ICTV designations.

