Family: Inoviridae
Petar Knezevic and Evelien M. Adriaenssens
The citation for this ICTV Report chapter is the summary published as Knezevic et al., (2021):
ICTV Virus Taxonomy Profile: Inoviridae, Journal of General Virology 2021 102(7):001614.
Corresponding author: Petar Knezevic (petar.knezevic@dbe.uns.ac.rs)
Edited by: Evelien M. Adriaenssens and Stuart G. Siddell
Posted: May 2021
PDF: ICTV_Inoviridae.pdf
Summary
Members of the family Inoviridae have virions with non-enveloped flexible filaments (600−2,500 × 6−10 nm) with supercoiled, circular, positive-sense single-stranded DNA [(+) ssDNA] genomes of about 5.5−10.6 kb, encoding 7−15 proteins (Table 1. Inoviridae). They infect Gram-negative bacteria and adsorb to bacterial pili, replicating their DNA genome by a rolling-circle mechanism, with progeny virions released from cells by extrusion without killing the host. Phage DNA can persist extra-chromosomally or integrate into the bacterial genome.
Table 1. Inoviridae. Characteristics of members of the family Inoviridae
| Characteristic | Description |
| Example | Escherichia phage M13 (V00604), species Inovirus M13 |
| Virion | Non-enveloped flexible filaments; 6−10 nm in diameter, 600−2,500 nm in length |
| Genome | 5.5−10.6 kb, supercoiled circular positive-sense single-stranded DNA, encoding 7−15 proteins |
| Replication | Rolling-circle mechanism |
| Translation | From mRNAs |
| Host range | Gram-negative bacteria |
| Taxonomy | Realm Monodnaviria, kingdom Loebvirae, phylum Hofneiviricota, class Faserviricetes, order Tubulavirales; the family Inoviridae includes 26 genera and 52 species |
Virion
Morphology
Virions of members of the family are characterized by a relatively simple morphology. Their protein coat is helically organized around a positive-sense single-stranded circular DNA. Virions are non-enveloped, long and flexible filaments. For members of the most widely studied species, Escherichia virus M13, represented by the exemplar isolate Escherichia phage M13 (M13), virions are built up from a major coat protein (CoaB; p8) present in 2700 copies, and four additional proteins present in only five copies: p7 and p9 at the blunt end and p3 and p6 at the rounded end [reviewed in (Rasched and Oberer 1986)] (Figure 1. Inoviridae).
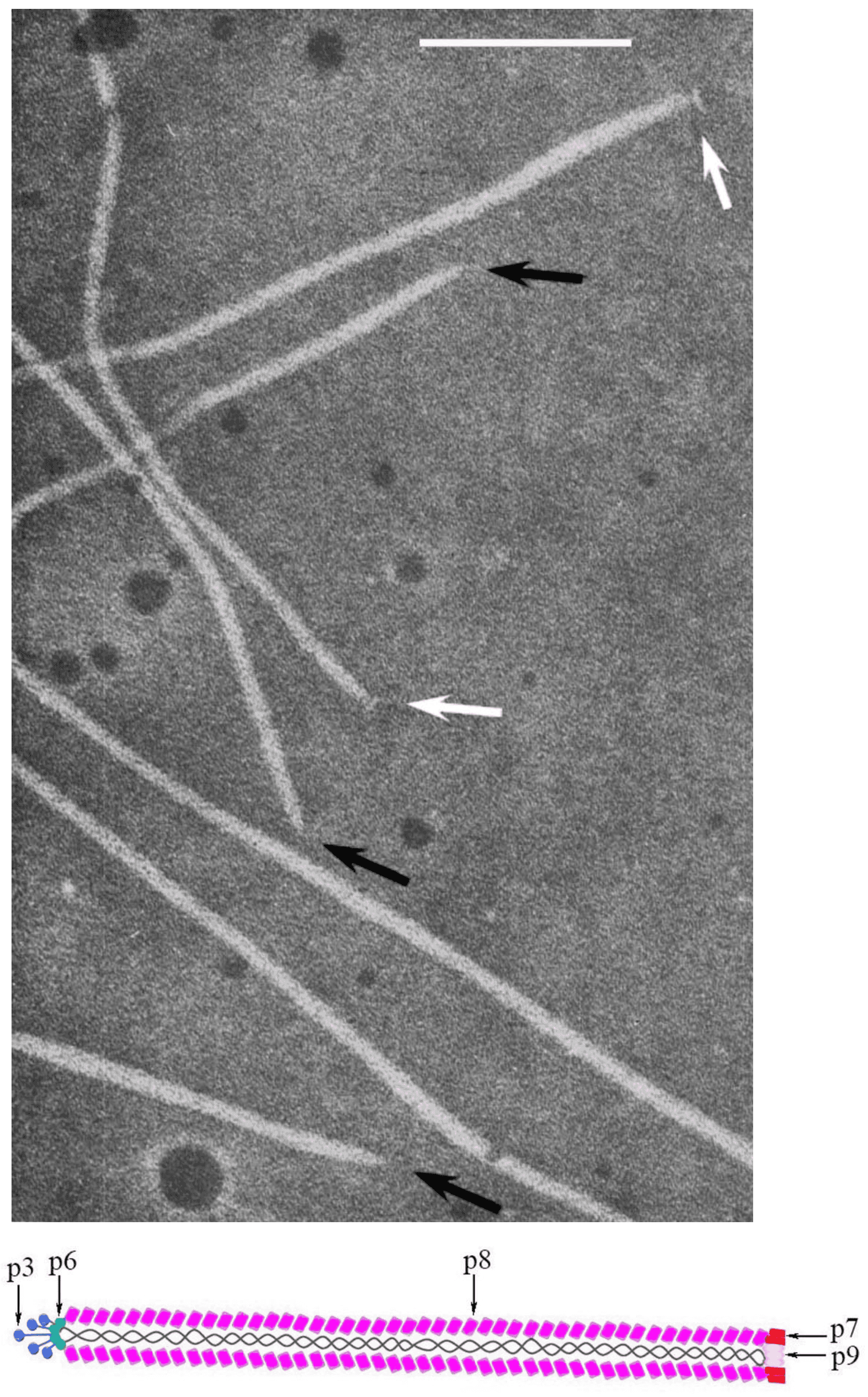 |
| Figure 1. Inoviridae. Virion structure of members of family Inoviridae: (Top) Negative-contrast electron micrograph shows a filamentous virion structure of a phage infecting enterobacteria; black arrows indicate pointed virion ends, while white indicate blunt ends. The bar represents 100nm (modified with permission from (Bradley et al., 1982); (Bottom) Schematic diagram of the virion Escherichia phage M13 (species Escherichia virus M13) |
The major coat protein is highly α-helical and its N-terminal domains are negatively-charged, hydrophilic and oriented outside the virion; hydrophobic central domains stabilize subunit–subunit interactions, while C-terminal domains interact electrostatically with the DNA (Lee et al., 2012). The interaction of DNA and proteins is established among phosphate groups of the DNA and residues of basic amino acids in the major coat protein. The mass ratio of proteins and DNA in the particle determines specific symmetry of the phages. There are two classes according to this criterion: class I comprising a 5-fold rotation axis combined with 2-fold screw axis (e.g. M13) and class II with a simple one-start helix with 5.4 subunits per turn (e.g. Pseudomonas phage Pf1 [Pf1]) (Marvin et al., 1994).
Virion length depends on both DNA length and the average nucleotide rise (h) which ranges from 0.24 nm in Pseudomonas phage Pf3 and 0.28 nm in M13 to 0.62 nm in Pseudomonas phage Pf4. Thus, the dimensions of virions varies from a length of 600 nm for Xanthomonas phage XacF1 to more than 2,500 nm for Vibrio phage VfO3K6, while the diameter varies from 6 nm for Vibrio phage VfO4K68 to 10 nm for Ralstonia phage RSM1 and Ralstonia phage RSS1. In contrast to icosahedral phages, there is no theoretical limit to the amount of DNA that can be packed, so short “microphage” containing less DNA (Specthrie et al., 1992) and “polyphage”, containing 10−20 DNA molecules packaged end-to-end into a single particle can also be produced (Lopez and Webster 1983).
Physicochemical and physical properties
The total mass of virions ranges from 10 MDa to over 60 MDa, while S20,w values are in a narrow range 40–45S and mass-per-length values are near 18,000 Da nm−1 (Day 2012). Virions can be precipitated with low concentration of polyethylene glycol 8000 (PEG8000) and can be purified using CsCl gradient ultracentrifugation in which their buoyant density is approx. 1.3–1.4 g cm−3.
Although virions do not contain lipids, they are sensitive to organic solvents. Sensitivity to diethyl ether, acetone, chloroform and methanol varies depending on the phage isolate: Pf1 is inactivated by 50% in these solvents for 10 min at room temperature, whereas Escherichia phage fd retains full infectivity in all of the above solvents except chloroform (Amako and Yasunaka 1977). The interaction of M13 filamentous virions with chloroform/water interface induces a morphological change, leading sequentially to contraction of the filaments into shortened rods (I-forms), and then into spheroidal particles (S-forms) (Oh et al., 1999). Inactivation of filamentous phages by methanol, ethanol and 1-propanol is inversely dependent upon alcohol hydrophobicity (Olofsson et al., 2001).
M13 has been confirmed as highly desiccation-resistant, exhibiting a half-life of up to 120 days. M13 is completely inactivated by strong acidic and alkaline conditions, but stable at pH 3−11 for 20 minutes. Incubation at 80 °C for 60 min decreases the titer by less than 1 log, while particles are completely inactivated after 50 min at 95 °C or after 20 min at 99 °C. Virion sensitivity to detergents and disinfectants is variable (Branston et al., 2013).
Nucleic acid
The genome is a positive-sense, supercoiled, circular ssDNA molecule of 5.5 to 10.6 kb, encoding 7−15 proteins. GC content is dependent on the host, varying from 40.5% for Salmonella phage Ike to 60.7% for Ralstonia phage PE226. Nucleic acid constitutes 6−14% of the virion weight.
Proteins
The coat of M13 is composed of 2,700 copies of the major coat protein (p8, CoaB), together with five copies of each of the minor coat proteins: p7 and p9 form a blunt assembly at the end of the filament while p3 (CoaA) and p6 form a rounded structure at the adsorption end. Structural proteins possess signal peptides for integration into cytoplasmic membrane before extrusion.
Lipids
Not reported.
Carbohydrates
Not reported.
Genome organization and replication
The positive-sense, circular ssDNA genome of members of the family Inoviridae is organized in modules (Figure 2. Inoviridae). The M13 genome has a DNA replication module (g2, g5 and g11), a structural module (g7, g9, g8, g3 and g6), and a morphogenesis, i.e. an assembly/extrusion module (g1, g4 and g11). Intergenic regions contain the origin of replication, packaging signals and promoters of various strengths.
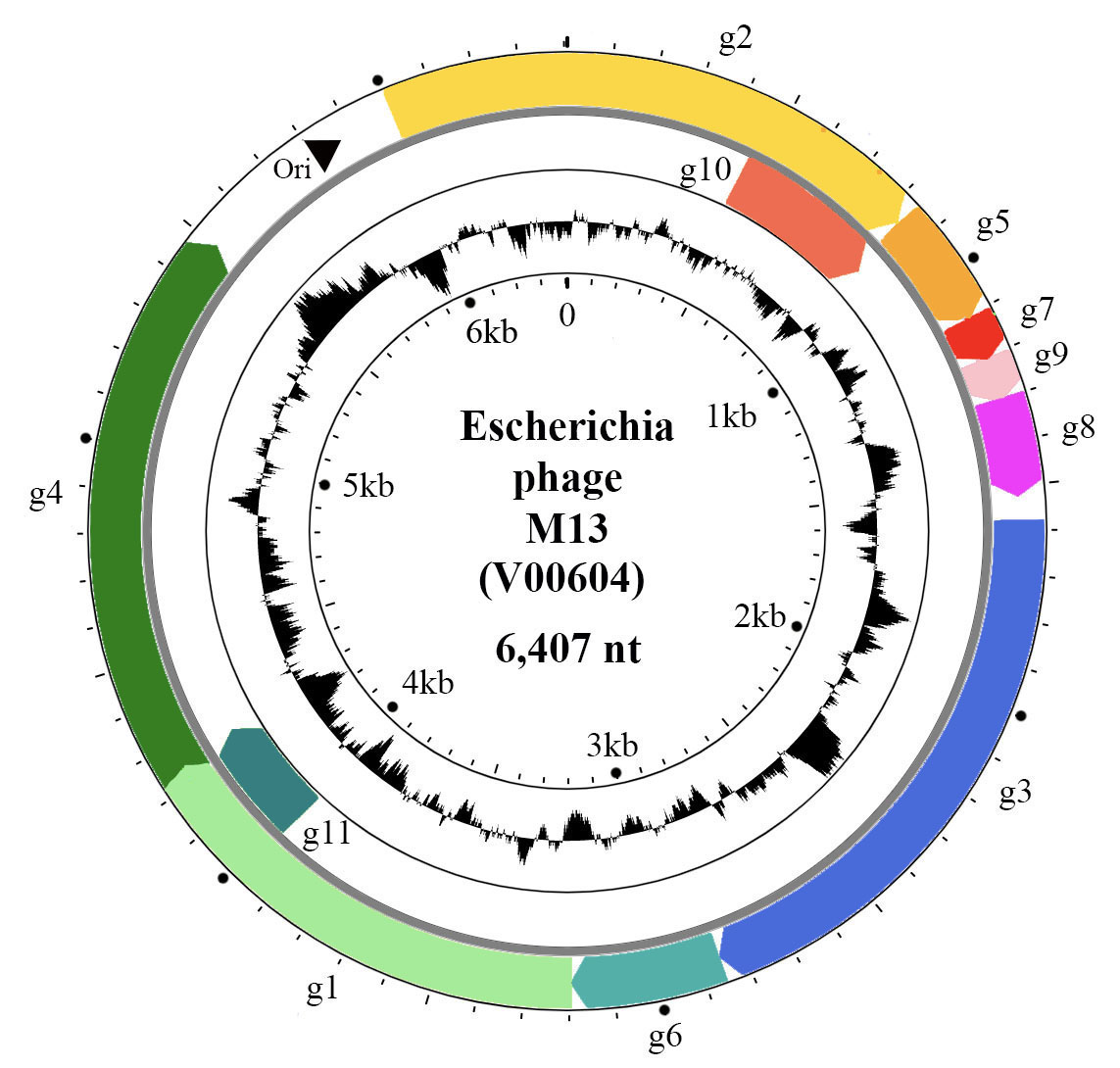 |
| Figure 2. Inoviridae. Circular representation of the Escherichia phage M13 genome. Genes (coloured arrows) are organised in modules of replication genes (g2, g5 and g10), structural genes (g7, g9, g8, g3, g6) and morphogenesis genes (g1, g4 and g11). Inner circle indicates GC content. |
The genes are densely packed on the genome and some of them overlap or are entirely within larger genes, being translated from internal start codons. For instance, g11 and g10 of M13 are C-terminal portions of g1 and g2, respectively (Figure 2. Inoviridae). Phages that are able to integrate into the bacterial chromosome have genomes containing insertion sequences and encode proteins for latent stage regulation (repressor); some phages encode integration proteins (integrase or transposase). Some genes, in particular the repressor genes of integrative phages, are expressed from the negative-sense strand, such as rstR gene of Vibrio phage CTXphi. The genomes of many bacteriophages also have accessory genes.
Inoviruses encode 7 to 15 proteins. The most widely studied inovirus is M13, for which the function of all 11 proteins is known, five of which are structural (Table 2. Inoviridae).
Table 2.Inoviridae. Escherichia phage M13 proteins
| Protein | Mol. mass (kDa) | Function |
| p2 | 46.0 | Replication initiation protein; endonuclease |
| p10 | 12.0 | A C-terminal fragment of p2 translated from an internal start codon; required for the accumulation of genomic (+) ssDNA |
| p5 | 9.8 | Helix destabilizing protein, with affinity to (+) ssDNA; it binds as homodimer to (+) ssDNA, preventing further conversion to replicative form (RF) |
| p7 | 3.5 | Minor coat protein; five copies at the tip of the virion that first emerges from the host cell |
| p9 | 3.3 | Minor coat protein; five copies at the tip of the virion that first emerges from the host cell |
| p8 | 5.4 | Major coat protein (CoaB); combined with ssDNA genome it forms the elongated filamentous particles |
| p3 | 43.0
| Minor coat protein (CoaA) located at opposite side of filament from p7 and p9; C-terminus helps terminate assembly, while the N-terminus attaches to pili and initiates infection |
| p6 | 12.0 | Minor coat protein located at the end of the virion along with gp3; responsible for the termination of virion assembly; when absent, phage particles remain associated with the host membrane |
| p1 | 35.0 | Morphogenesis protein (Zot; zonula occludens toxin) with ATPase function; an inner membrane protein that interacts with the outer membrane protein p11 |
| p11 | 8.0 | C-terminal fragment of p1, translated from an internal start codon; interacts with p1, being involved in extrusion |
| p4 | 50.0 | Forms a homo-multimeric channel (14-mer) in membrane for phage secretion; along with p1/p11 forms assembly machinery. |
Among inoviruses, the replication cycle of M13 has been studied in most detail (Figure 3. Inoviridae). Infection starts when p3 interacts with the tip of a pilus on the bacterial surface (adhesion receptor) (1). This interaction triggers pilus retraction (2), bringing the phage closer to the bacterial cell surface, in proximity of TolQRA co-receptor (entry receptor) (Click and Webster 1998) (3). The phage (+) ssDNA genome is subsequently translocated into the bacterial cytoplasm and the capsid proteins are integrated into the host cytoplasmic membrane (4). The (+) ssDNA phage genome is converted to covalently bound dsDNA, the replicative form (RF), using a short primer generated by host RNA polymerase and the complementary strand of (-) ssDNA synthesized by activity of host DNA polymerase III (Geider and Kornberg 1974) (5). mRNAs are transcribed from the RF by the host RNA polymerase (6) and further translated by host machinery to produce the phage proteins (7). Translation involves the use of overlapping reading frames and alternate starts in the same frame. Replication of DNA starts when p2, which possesses endonuclease activity, nicks dsDNA at the origin of replication (Meyer et al., 1979) (8). The host polymerase then synthesizes (+) ssDNA from the complementary strand of RF by a rolling circle mechanism (9). The generated (+) ssDNA molecules are further converted into RFs (10) which are also used for transcription and more phage proteins are formed. Translated p5 has affinity to ssDNA and when its concentration is high enough, homodimers cover newly synthesized (+) ssDNA molecules (11). This inhibits further conversion of ssDNA into RFs and collapses DNA into filaments with an uncovered double stranded packaging signal at one end of the filament (12). The corresponding structural proteins integrate into the cytoplasmic membrane (13) along with the assembly proteins, these being a 14-mer aggregate of p4, plugged with p1 and p11 forming a pore for virion extrusion through the inner and outer membrane (Feng et al., 1999) (14). Virions are released by extrusion during the replacement of p5 with p8, the major coat protein (15). Proteins p7 and p9 are the first that extrude, followed by numerous copies of p8, while p3 and p6 are added at the end of the process and new virions are released into environment (16).
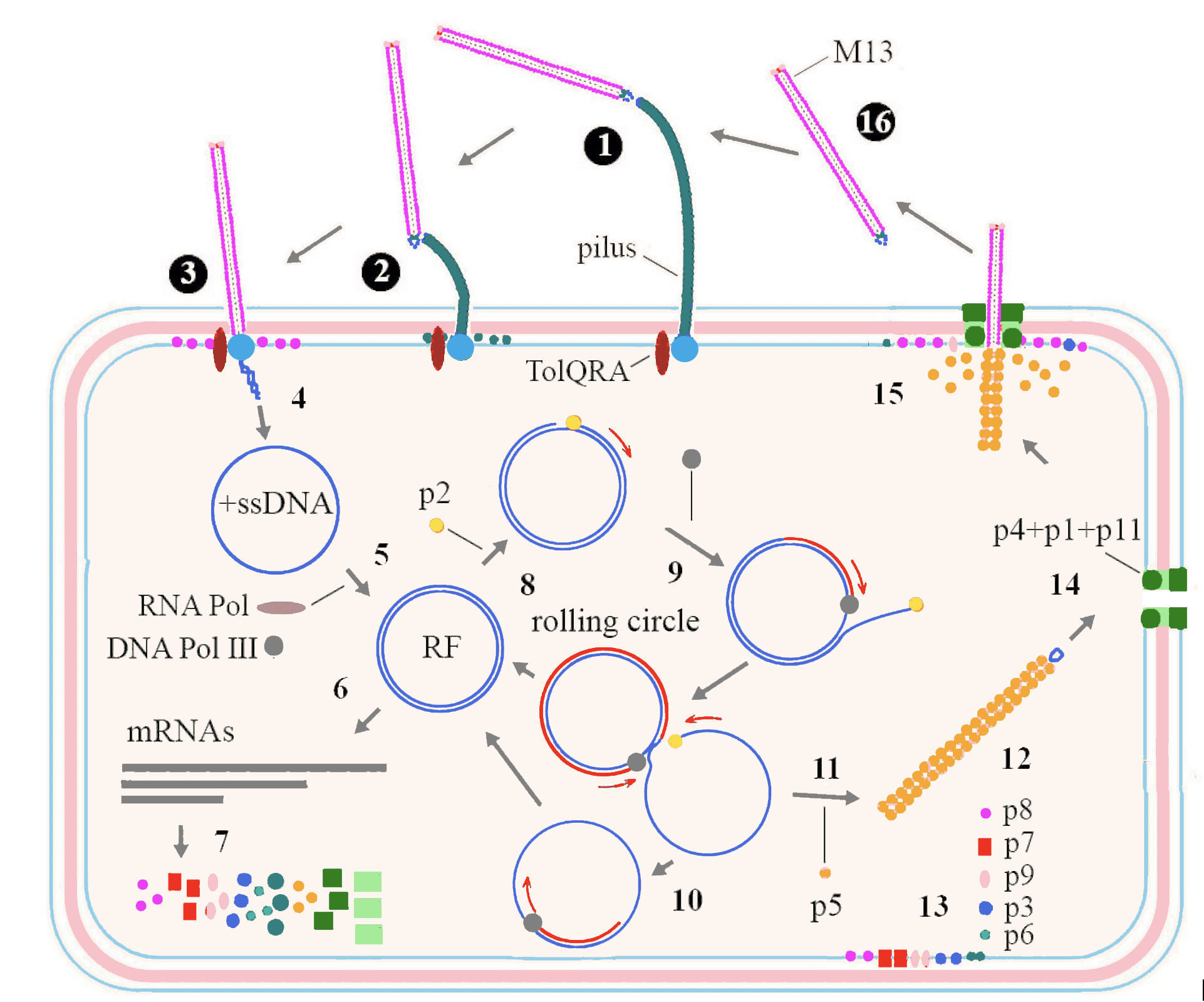 |
| Figure 3. Inoviridae. Replication cycle of a typical member of the family Inoviridae. Details are in the text above. |
Biology
Hosts for phages belonging to the family Inoviridae are Gram-negative bacteria and so far phages infecting the following genera have been described: Escherichia, Salmonella, Pseudomonas, Xanthomonas, Stenotrophomonas, Vibrio and Ralstonia. The receptors for some phages are conjugative pili encoded by plasmids which can be transferred by lateral gene transfer to new bacterial strains, so making them phage-sensitive. Other types of pili can also be utilized as receptors, such as type IV pili which are responsible for twitching motility.
Members of the family are neither obligatory lytic nor typically temperate phages, and enter neither the lytic nor typical lysogenic cycle. Inovirids establish a chronic productive infection, continuously releasing virions from infected cells by extrusion while the cells stay viable and intact. The infected cell usually exhibits a slower growth rate, and many inovirids do not produce plaques, or if they do, the plaques are usually turbid.
The DNA of some inovirids persists in the bacterial cell in a circular, extra-chromosomal form similar to plasmids, being unable to integrate into the cell genome. Other inovirids can integrate into the bacterial chromosome by means of a phage-encoded site-specific integrase or transposase. Inovirids that possess an integrase usually integrate at a specific site in the bacterial genome, mainly in tRNA genes, while those phages with transposases are able to integrate at several sites. In some cases, phage DNA is integrated into the bacterial genome although the phage does not encode any recognizable integration enzymes. In this case corresponding host enzymes are involved into the process, such as V. cholerae XerC/D recombinase that mediates integration at the chromosome dimer resolution site dif (Huber and Waldor 2002, McLeod et al., 2005). The mechanisms of prophage persistence vary significantly, and in Vibrio phage CTXphi the phage protein RstR and the host-encoded LexA are involved in latency. Mechanisms have not been described for processes such as insertion, repression, immunity, induction and involvement in transduction.
Prophages of members of the family Inoviridae can be detected in many bacterial genomes (Roux et al., 2019); these prophages are not assigned to virus species. For instance, more than 60% of P. aeruginosa strains possess at least one Pseudomonas phage Pf1-related genetic element (Knezevic et al., 2015). Usually more than one prophage can be detected in a bacterial strain, and sometimes they are organized as tandem repeats of two to four copies, such as Yersinia prophage Ypfphi of Y. pestis or Vibrio phage CTXphi of V. cholera (Bischerour et al., 2012, Derbise and Carniel 2014). This suggests that these phages may play a role in bacterial evolution and variability.
Some phages encode genes that are involved in bacterial virulence and upon integration into the bacterial genome cause lysogenic conversion of their hosts. A typical example is Vibrio phage CTXphi that encodes three cholera-related toxins: CT - cholera toxin, Zot - zonula occludens toxin and Ace - accessory cholera toxin; the role of phage in V. cholerae virulence has been well documented (Fasano et al., 1991, Trucksis et al., 1993, Waldor and Mekalanos 1996). In contrast, some phages can decrease virulence, such as the interaction of members of the genus Habenivirus with the plant pathogen Ralstonia solanacearum (Addy et al., 2012). Inovirus involvement in bacterial virulence has been postulated for many other plant, human and animal pathogens (e.g. Yersiania pestis, Neisseria meningitidis, Pseudomonas aeruginosa), affecting biofilm formation, exopolisaccharide production, various types of motility etc. (Bille et al., 2005, Gonzalez et al., 2002, Webb et al., 2004). Similarly, inovirids contribute to the fitness of non-pathogenic bacteria; for instance, an unclassified filamentous phage enhances motility and chemotaxis of Pseudoalteromonas spp., contributing to host survival in Arctic sea ice (Yu et al., 2015).
The filamentous phages, particularly Escherichia phage M13, have been extensively used as cloning vectors (phagemids) in DNA sequencing, phage display technology, and in other domains of biotechnology and nanotechnology.
Genus demarcation criteria
For genus demarcation, beside considerable similarity of DNA sequences confirmed by BLASTn, the phage should have significant similarity of both Zot and Coat B (major capsid) proteins. Each of the proposed genera comprise phages with similar DNA sequences; phages in different genera differ from each other by >50% in the amino-acid sequence of the major coat (CoaB; p8) and morphogenesis (Zot; p1) proteins as assessed using the BLASTp algorithm.
Species demarcation criteria
Members of the same species are >95% identical in genome DNA sequence and show significant similarity (identity × query coverage) in their adhesion protein (CoaA; p3) amino acid sequences.
Derivation of names
Most genus names in the family are derived from Latin words for or relating to long, thin objects, particularly those associated with weaving, referring to the filamentous nature of phage particles.
Affertcholeramvirus-: from the Latin affero (third person singular in present tense affert) - meaning “it brings” and the Latin cholera, cholerae – meaning the disease cholera.
Capistrivirus: from the Latin capistrum, capistri meaning a halter.
Coriovirus: from the Latin corius, cori(i) meaning a thong.
Fibrovirus: from the Latin fibra, fibrae meaning a fiber.
Habenivirus: from the Latin habena, habenae meaning a strap.
Infulavirus: from the Latin infula, infulae meaning a woolen headband knotted with ribbons.
Inoviridae, Inovirus: from the Greek is, inos (ἴς, ἰνός) meaning a fiber.
Lineavirus: from the Latin linea, lineae meaning line.
Parhipatevirus: from the Latin parhypate, parhypatus meaning the second-top string/note or next to highest
Primolicivirus: from the Latin primus meaning first and the Latin licium, licii – meaning a thread.
Psecadovirus: from the Latin psecas, psecadis meaning an anointer of hair
Restivirus: from the Latin restis, restis meaning a rope.
Saetivirus: from the Latin saeta, saetae meaning horse hair.
Scuticavirus: from the Latin scutica, scuticae meaning a lash.
Staminivirus: from the Latin stamen, staminis meaning the warp of a loom.
Subteminivirus: from the Latin subtemen, subtemini meaning the woof in weaving.
Tertilicivirus: from the Latin words tertius meaning third and licium, licii meaning a thread.
Versovirus: from the Latin versus, versus meaning a line.
Vicialiavirus: from the Latin vicialia, vicialium meaning stalks.
Villovirus: from the Latin villus, villi meaning shaggy hair.
Xylivirus: from the Latin xylon, xyli meaning cotton.
Relationships within the family
The morphogenesis protein (p1 or Zot) is conserved in inoviruses and phylogenetic relationships within the family, based on the amino-acid sequence of this protein, are depicted in Figure 4. Inoviridae. There are 2 major clades in the Inoviridae family tree and each clade bifurcates into subclades, all supported by high bootstrap values. Although a relatively small number of species and genera have been described, numerous prophage similar to members of the Inoviridae have also been detected in bacterial genomes.
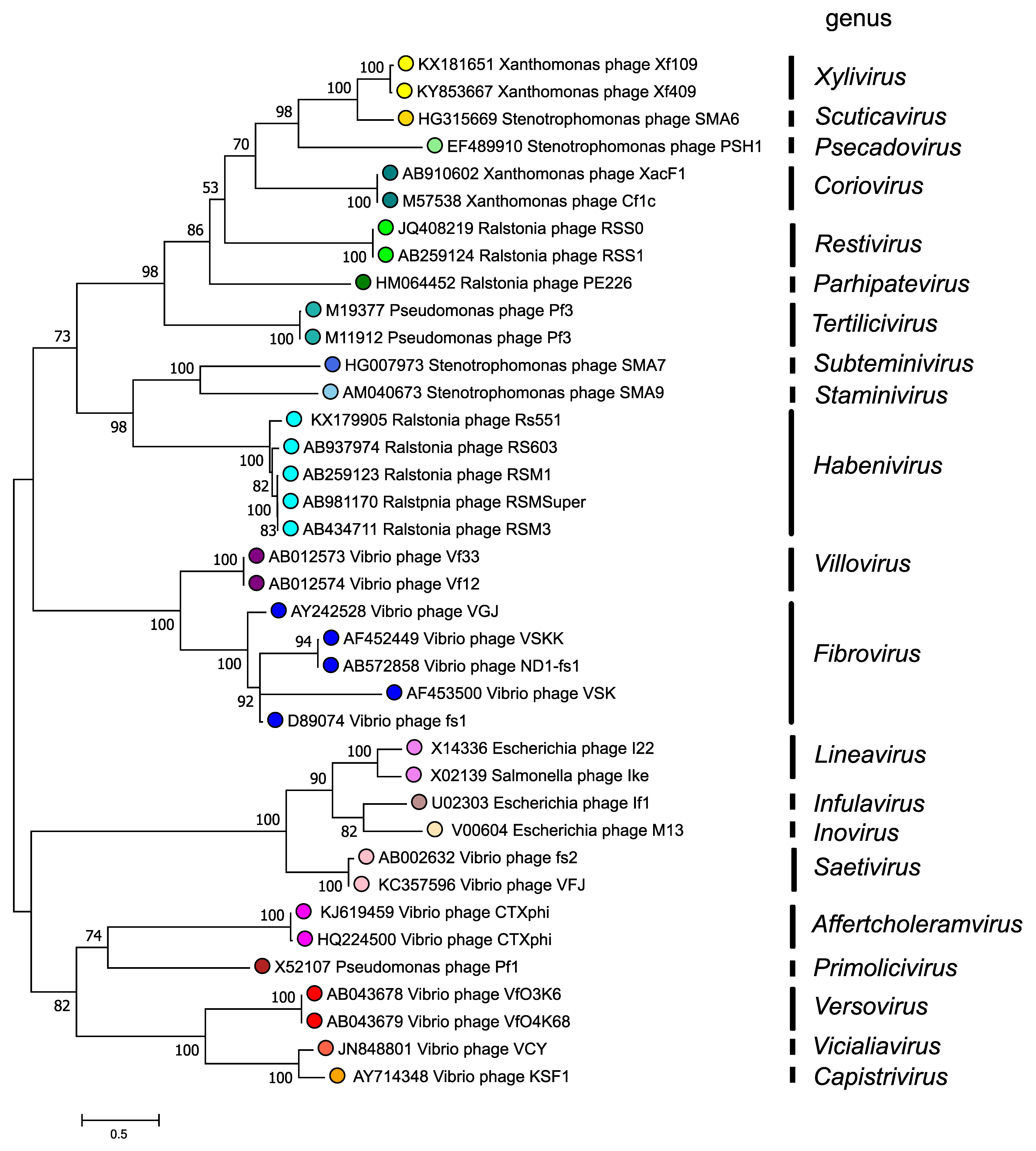 |
| Figure 4. Inoviridae. Phylogenetic relations of members of the family Inoviridae. Maximum Likelihood phylogenetic tree inferred using the Whelan And Goldman + Freq. model (Whelan and Goldman 2001) from a MUSCLE alignment of the full-length p1 (Zonula occludens toxin) sequences of 40 inovirids assigned to 21 genera. The tree with the highest log likelihood (-20779.30) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (2 categories (+G, parameter = 6.0712)). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 38 amino acid sequences. The percentage of 100 trees in which the associated taxa clustered together is shown next to the branches where this was > 50%. There were a total of 603 positions in the final dataset. The analyses were conducted in MEGA X (Kumar et al., 2018, Stecher et al., 2020). |
Relationships with other taxa
Members of the family Inoviridae share many similar features with those of the families Plectroviridae and Paulinoviridae. All three families are included in the order Tubulavirales, comprising viruses of prokaryotes with helical symmetry, a (+) ssDNA genome and that establish a chronic productive infection with virions released by extrusion.

