Family: Marnaviridae
Andrew S. Lang, Marli Vlok, Alexander I. Culley, Curtis A. Suttle, Yoshitake Takao and Yuji Tomaru
The citation for this ICTV Report chapter is the summary published as Lang et al., (2021):
ICTV Virus Taxonomy Profile: Marnaviridae 2021, Journal of General Virology, 2021 102(8):001633.
Corresponding author: Andrew S. Lang ([email protected])
Edited by: Sead Sabanadovic and Stuart G. Siddell
Posted: June 2021, updated May 2025
Summary
Marnaviridae is a family of small non-enveloped viruses with positive-sense RNA genomes of 8.6–9.6 kb (Table 1 Marnaviridae). Isolates infect marine single-celled eukaryotes (protists) that come from diverse lineages. Also, some members are known from metagenomic studies of ocean virioplankton, with additional unclassified viruses described from metagenomic datasets from marine and freshwater environments.
Table 1 Marnaviridae. Characteristics of members of the family Marnaviridae
| Characteristic | Description |
| Example | Heterosigma akashiwo RNA virus (AY337486), species Marnavirus taichanarum |
| Virion | Non-enveloped, 22–35 nm virions with four structural proteins |
| Genome | 8.6–9.6 kb of positive-sense, non-segmented RNA |
| Replication | Cytoplasmic, involving RNA-directed RNA polymerase and helicase; cytolytic |
| Translation | Directly from genomic RNA containing one or more internal ribosomal entry sites (IRES) |
| Host range | Single-celled eukaryotes (protists) from marine environments |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Pisuviricota, class Pisoniviricetes, order Picornavirales; 7 genera and 20 species |
Virion
Morphology
Those members of the family Marnaviridae that have been isolated and structurally characterized have polyhedral virions that range from 22 to 35 nm in diameter, do not appear to have an envelope, and have no discernible projections (Figure 1 Marnaviridae; Table 2 Marnaviridae).
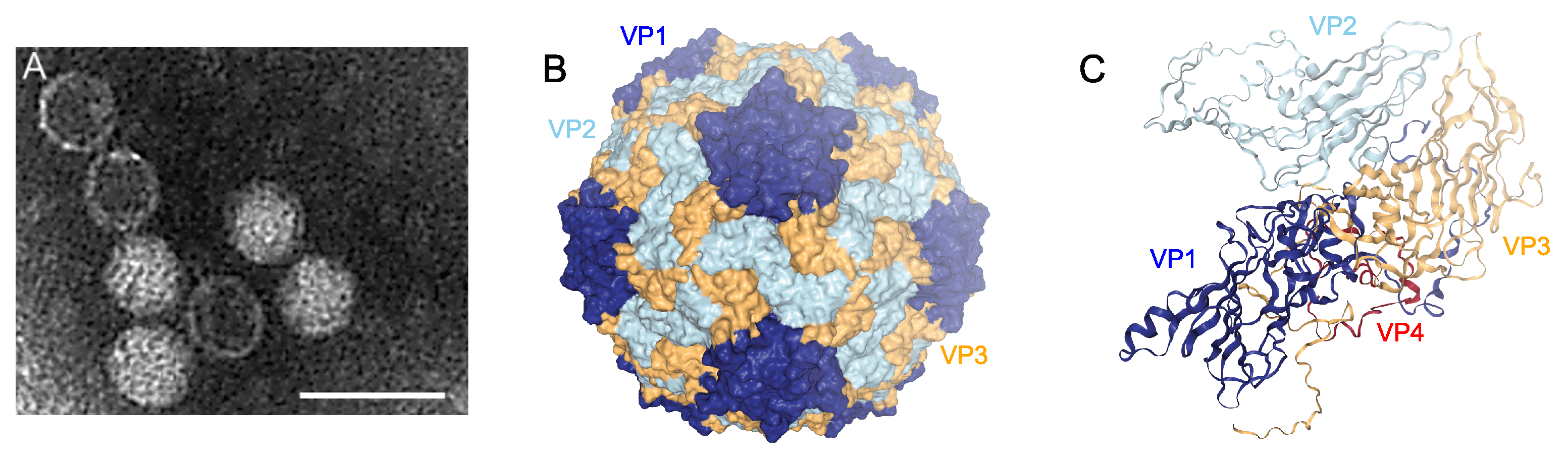 |
| Figure 1 Marnaviridae. virion structures. A. Negatively-stained transmission electron micrograph of Heterosigma akashiwo RNA virus (species Marnavirus taichanarum) particles (scale bar = 50 nm). B. Cryo-electron microscopy reconstructed capsid structure at 3.1 Å resolution of Chaetoceros tenuissimus RNA virus type II (species Sogarnavirus kimuraei) (PDB structure 6SHL viewed in NGL-WebGL). The three major capsid proteins are represented in dark blue (VP1), light blue (VP2) and yellow (VP3). C. Ribbon depiction of the protein subunits from the reconstruction in B, including the minor capsid protein VP4 (red). |
Physicochemical and physical properties
Virions are not sensitive to chloroform.
Nucleic acid
Members of the family Marnaviridae possess monopartite, positive-sense RNA genomes containing either one or two open reading frames (ORFs) (Figure 2 Marnaviridae). One exception is Aurantiochytrium single-stranded RNA virus 01 (AuRNAV01, species Labyrnavirus takaoii), which has an additional ORF at the 3′-end of the genome. Among the isolated viruses the genome lengths range from 8.6 to 9.6 kb including a 3′-poly(A) tail (Table 2 Marnaviridae). Genome lengths for viruses known only from metagenomic sequencing data fall within this range, although some of these genome sequences are not complete (Table 3. Marnaviridae). Predicted secondary structures within the 5′-non-coding regions (NCRs) and intergenic regions (IGRs) suggest the presence of internal ribosome entry sites (IRES) (Figure 2 Marnaviridae), a feature observed in other members of other families in the order Picornavirales.
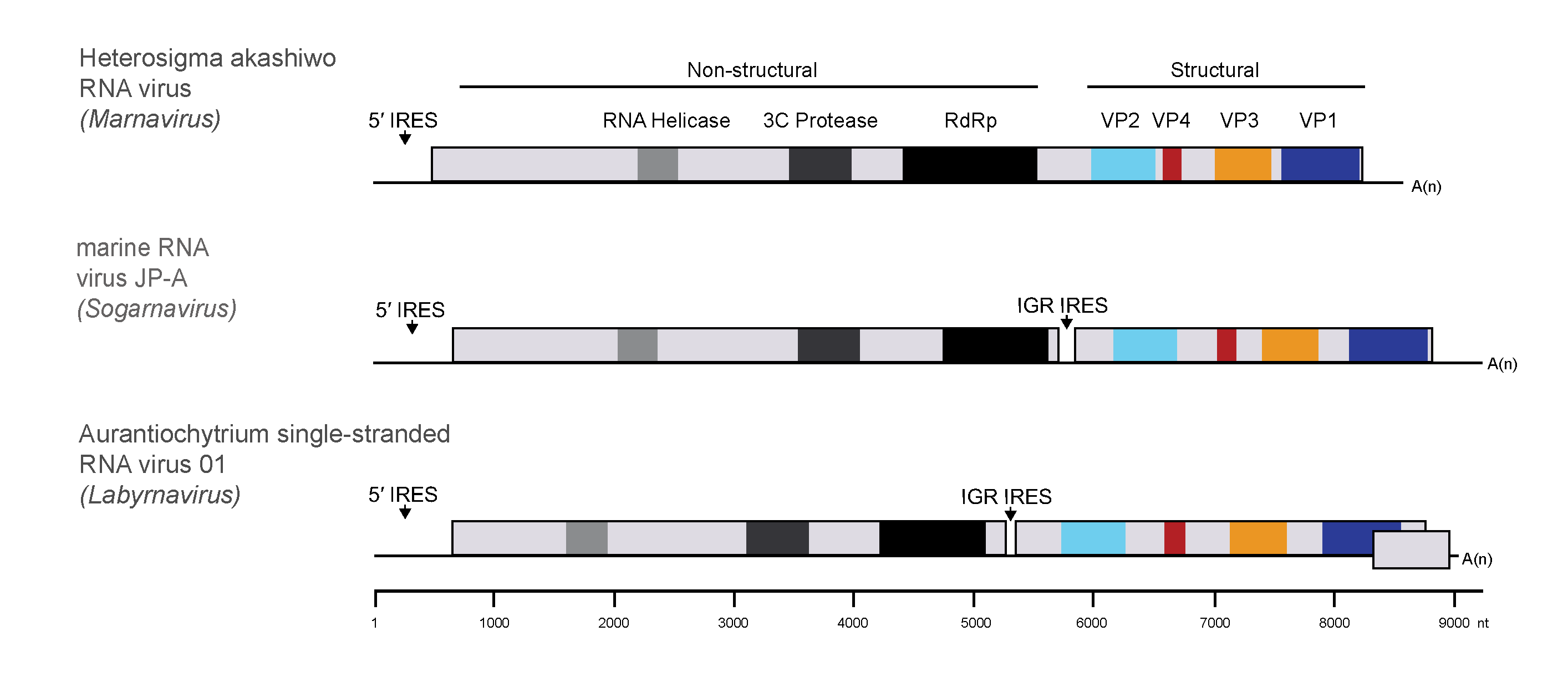 |
| Figure 2 Marnaviridae. Genome architecture of members of the family Marnaviridae. All genomes are organized with the non-structural proteins, including RNA helicase, 3C protease and RNA-directed RNA polymerase domains, at the 5′-ends. The structural proteins (VP2, VP4, VP3 and VP1) are encoded at the 3′-ends of the genomes, downstream of a putative internal ribosome entry site in the intergenic region in the di-cistronic genomes. |
Proteins
Members of the family Marnaviridae encode four conserved proteins, VP1–4, with recognizable similarities to the structural proteins of other members of the order Picornavirales. The three largest of these (VP1–3) have been visualized by SDS-PAGE for some isolates (Tai et al., 2003, Shirai et al., 2008, Nagasaki et al., 2004). The structure of all four capsid proteins as well as the virion structure have been resolved by cryo-electron microscopy for Chaetoceros tenuissimus RNA virus type II (species Sogarnavirus kimuraei) (Munke et al., 2020) (Figure 1 Marnaviridae).
Table 2 Marnaviridae. Properties of Marnaviridae isolates
| Genus | Species | Virus name (abbreviation) | Host species (type)a | Virion diameter (nm) | Genome length (kb) | Latent period (hours) | Burst size | References |
| Marnavirus | Marnavirus taichanarum | Heterosigma akashiwo RNA virus (HaRNAV) | Heterosigma akashiwo (R) | 25 | 8.6 | 48 | 460–520 | (Tai et al., 2003, Lang et al., 2004) |
| Bacillarnavirus | Bacillarnavirus nagasakii | Rhizosolenia setigera RNA virus 01 (RsRNAV01) | Rhizosolenia setigera (D) | 32 | 8.9 | 48 | 1.0–3.1×103 | (Nagasaki et al., 2004, Shirai et al., 2006) |
| Bacillarnavirus | Bacillarnavirus setoensis | Chaetoceros tenuissimus RNA virus 01 (CtenRNAV01) | Chaetoceros tenuissimus (D) | 31 | 9.4 | <24 | 1.0×105 | (Shirai et al., 2008) |
| Bacillarnavirus | Bacillarnavirus yujii | Chaetoceros socialis f. radians RNA virus 01 (CsfrRNAV01) | Chaetoceros socialis f. radians (D) | 22 | 9.5 | <48 | 66 | (Tomaru et al., 2009) |
| Sogarnavirus | Sogarnavirus kimuraei | Chaetoceros tenuissimus RNA virus type II (CtenRNAVII) | Chaetoceros tenuissimus (D) | 35 | 9.6 | 24–48 | 287 | (Kimura and Tomaru 2015) |
| Sogarnavirus | Sogarnavirus tomaruii | Chaetoceros species RNA virus 02 (CspRNAV02) | Chaetoceros sp. (D) | 32 | 9.4 | <48 | NDb | (Tomaru et al., 2013) |
| Labyrnavirus | Labyrnavirus takaoii | Aurantiochytrium single-stranded RNA virus 01 (AuRNAV01) | Aurantiochytrium (T) | 25 | 9.0 | 8 | 0.58–6.4×104 | (Takao et al., 2006, Takao et al., 2005) |
| Kusarnavirus | Kusarnavirus tomaruii | Asterionellopsis glacialis RNA virus (AglaRNAV) | Asterionellopsis glacialis (D) | 31 | 8.8 | 48 | ND | (Tomaru et al., 2012) |
a R, raphidophyte; D, diatom; T, thraustochytrid
b ND, not determined
Table 3 Marnaviridae. Marnaviridae members discovered by metagenomics.
| Genus | Species | Virus name (abbreviation) | Genome length (nt)a | Genome completenessb | Source | References |
| Locarnavirus | Locarnavirus jerichoensis | marine RNA virus JP-B (JP-B) | 8926 | CG | Coastal marine | (Culley et al., 2007) |
| Locarnavirus derisii | marine RNA virus SF-2 (SF-2) | [9321] | CCG | Coastal wastewater | (Greninger and DeRisi 2015) | |
| Locarnavirus greningerii | marine RNA virus SF-1 (SF-1) | [8970] | CCG | Coastal wastewater | (Greninger and DeRisi 2015) | |
| Locarnavirus rohweri | marine RNA virus SF-3 (SF-3) | [8648] | CCG | Coastal wastewater | (Greninger and DeRisi 2015) | |
| Salisharnavirus | Salisharnavirus britensis | marine RNA virus BC-4 (BC-4) | [8593] | CCG | Coastal/oceanic marine | (Vlok et al., 2019) |
| Salisharnavirus stewardii | marine RNA virus PAL473 (PAL473) | [6360] | CCG | Coastal marine | (Miranda et al., 2016) | |
| Salisharnavirus vlokiae | marine RNA virus BC-1 (BC-1) | [8638] | PG | Coastal marine | (Vlok et al., 2019) | |
| Salisharnavirus mirandaeae | marine RNA virus PAL128 (PAL128) | [8660] | CCG | Coastal marine | (Miranda et al., 2016) | |
| Sogarnavirus | Sogarnavirus gustavseniae | marine RNA virus BC-2 (BC-2) | [8843] | CCG | Coastal marine | (Vlok et al., 2019) |
| Sogarnavirus palmerensis | marine RNA virus PAL156 (PAL156) | [7897] | CCG | Coastal marine | (Miranda et al., 2016) | |
| Sogarnavirus kitsilanoensis | marine RNA virus BC-3 (BC-3) | [8496] | PG | Coastal marine | (Vlok et al., 2019) | |
| Sogarnavirus culleyi | marine RNA virus JP-A (JP-A) | 9236 | CG | Coastal marine | (Culley et al., 2007) |
a Brackets indicate the genome sequence is not complete
b CG, complete genome; CCG, complete coding genome, protein-coding region(s) complete; PG, partial genome, protein-coding region(s) not complete
Genome organization and replication
Among the members of the 20 species and 7 genera in the family, there is a mixture of mono- and di-cistronic genome organizations (Figure 2 Marnaviridae). The majority of viruses have a di-cistronic genome organization, whereas Heterosigma akashiwo RNA virus (HaRNAV, genus Marnavirus) and marine RNA virus SF-3 (SF-3, genus Locarnavirus) have a single predicted polyprotein encoded in their genomes. Aurantiochytrium single-stranded RNA virus 01 (AuRNAV01, genus Labyrnavirus) contains an additional ORF at the 3′-end of the genome, which is transcribed as a sub-genomic mRNA during infection (Takao et al., 2006). Maps of protein domain organization within the polyprotein sequences are shown in Figure 2 Marnaviridae. Domains were identified using profile hidden Markov models (HMMs) based on similarities with conserved domains from other members of the order Picornavirales, including helicase, RNA-directed RNA polymerase (RdRP), and structural (VP) domains. Some genomes of viruses in the family also contain regions that resemble the 3C cysteine proteinases of members of the family Picornaviridae. Regardless of whether the genomes are mono- or di-cistronic, the non-structural proteins are encoded in the 5′-region and the structural proteins are encoded in the 3′region. Members of family Marniviridae do not appear to encode virus protein genome-linked (VPg)-like proteins, characteristic of some members of the order Picornavirales.
Biology
Where known, virus hosts are marine, single-celled, eukaryotic microbes (protists) (Table 2 Marnaviridae). Replication is cytolytic, with latent periods ranging from 8 to 48 hours. Cytopathic effects have been observed, with ultrastructural changes including swelling of the endoplasmic reticulum, the formation of crystalline arrays in the cytoplasm, vacuolation and disintegration of the cytoplasm, and the appearance of fibrous material in vacuolated areas (Tai et al., 2003, Nagasaki et al., 2004, Tomaru et al., 2009, Takao et al., 2005, Tomaru et al., 2012, Tomaru et al., 2004) . Large numbers of viral particles have been observed in the cytoplasm of infected cells (Shirai et al., 2008, Nagasaki et al., 2004, Takao et al., 2005). Burst sizes of these viruses vary from 66 to 105 particles per cell (Table 2. Marnaviridae).
Derivation of names
Bacillarnavirus: from Bacillariophyceae, the class of diatoms that are hosts of these RNA viruses
Kusarnavirus: from the Afrikaans word kus meaning coastal and RNA virus
Labyrnavirus: from Labyrinthulid, the host name and RNA virus
Locarnavirus: from Locarno Beach where samples for the first marine RNA virus metagenomes in this genus were collected
Marnaviridae, Marnavirus: from Latin mare, meaning “sea”, and RNA virus
Salisharnavirus: from Salish Sea, the water mass around coastal southern British Columbia, Canada, from which the first marine RNA virus sequences in this genus were amplified
Sogarnavirus: from the Strait of Georgia, a major water body of the Salish Sea, the water mass around coastal southern British Columbia, Canada, from which the first marine RNA virus sequences in this genus were amplified
Genus demarcation criteria
Genera are defined based on phylogenetic relationships between virus amino acid sequences. Phylogenetic analysis of RdRP domain sequences place the viruses in seven clades that correspond to genera (Figure 3 Marnaviridae).
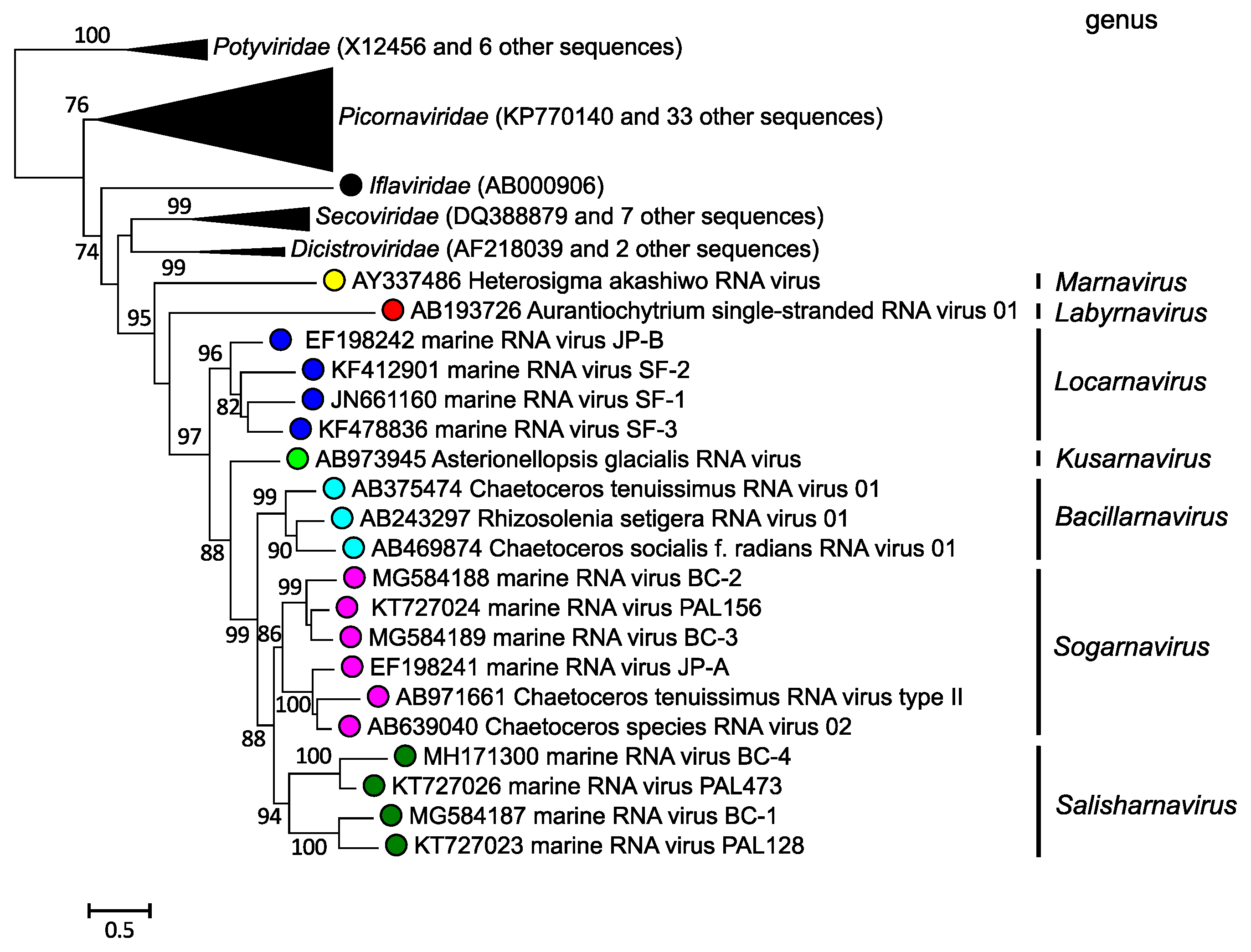 |
| Figure 3 Marnaviridae. Maximum likelihood phylogeny of RdRP amino acid sequences of members of the family Marniviridae. Branches containing sequences from viruses in other families in the order Picornavirales and the Potyviridae (outgroup) are collapsed. SH-like branch support values are indicated at the nodes when >0.70. The scale bar indicates average residue substitution per site. Modified from (Vlok et al., 2019). |
Species demarcation criteria
In genera with more than one species, species demarcation cut-offs are based on pairwise comparisons of the RdRP and capsid amino acid sequences. The cut-offs are 90% identity for the RdRP and 75% identity for the capsid.
Relationships within the family
Phylogenetic analysis of RdRP domain sequences place the 20 classified viruses in seven clades that correspond to genera (Figure 4 Marnaviridae, Figure 5 Marnaviridae).
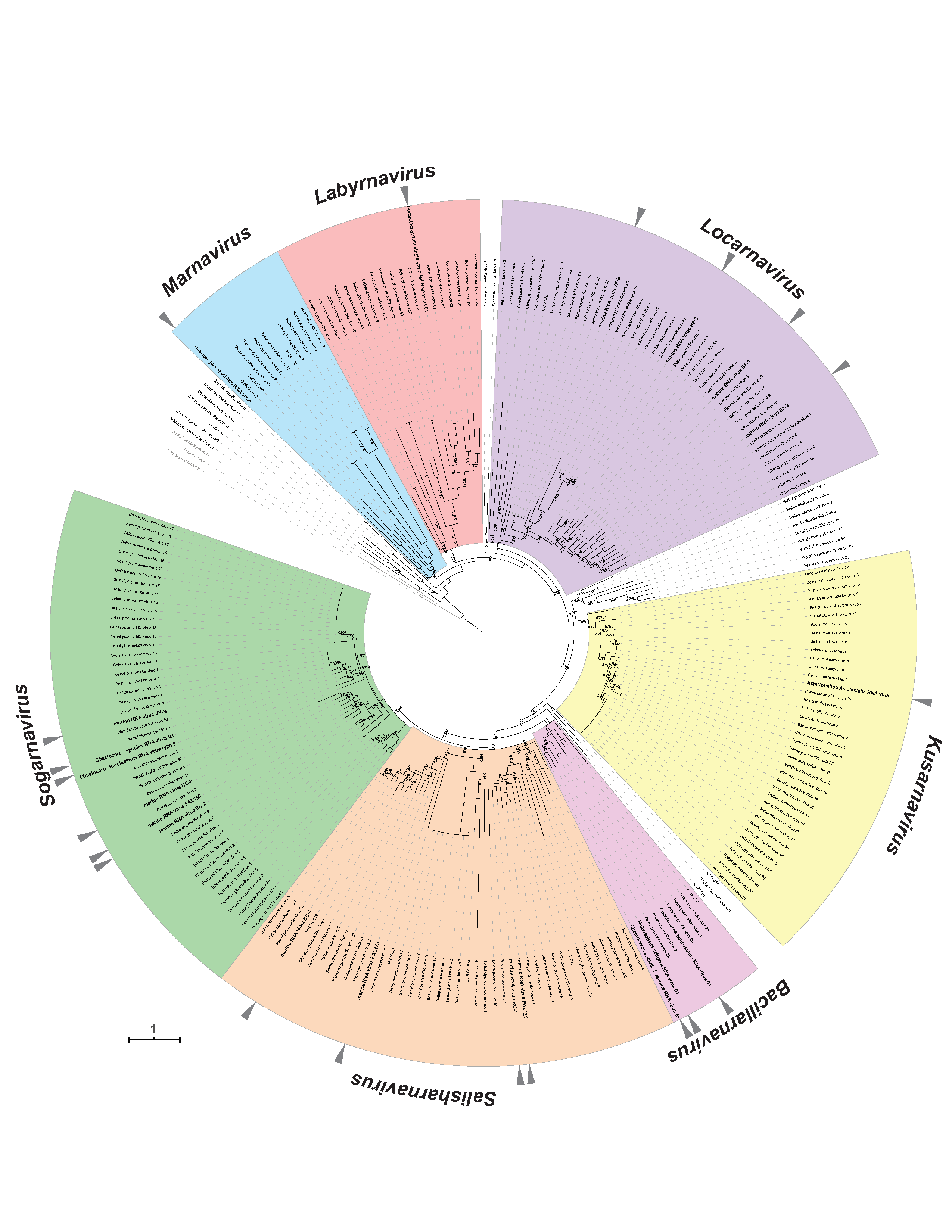 |
| Figure 4 Marnaviridae. RdRP phylogeny of viruses in the family Marnaviridae. Sequences outside of the clades of established genera are not highlighted in colour. Triangles indicate viruses classified into species. The RdRP conserved domains (Koonin and Dolja 1993) were aligned using MUSCLE v3.8.425 with default parameters (Edgar 2004) and manually curated with Aliview version 1.17.1 (Larsson 2014). The maximum-likelihood tree was constructed with PhyML 3.0 (Guindon et al., 2010) and the LG+I+G+F amino acid model (selected with Smart Model Selection (Lefort et al., 2017). Branch support was evaluated with the Shimodaira-Hasegawa approximate-likelihood ratio test and values are indicated at the nodes when >0.70. The maximum-likelihood scale bar indicates average residue substitution per site. The tree was edited in iTOL v3 (Letunic and Bork 2016). Accession numbers of individual viruses are given in the trees in the genera sections and in Figure 5. Marnaviridae. Modified from (Vlok et al., 2019). This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
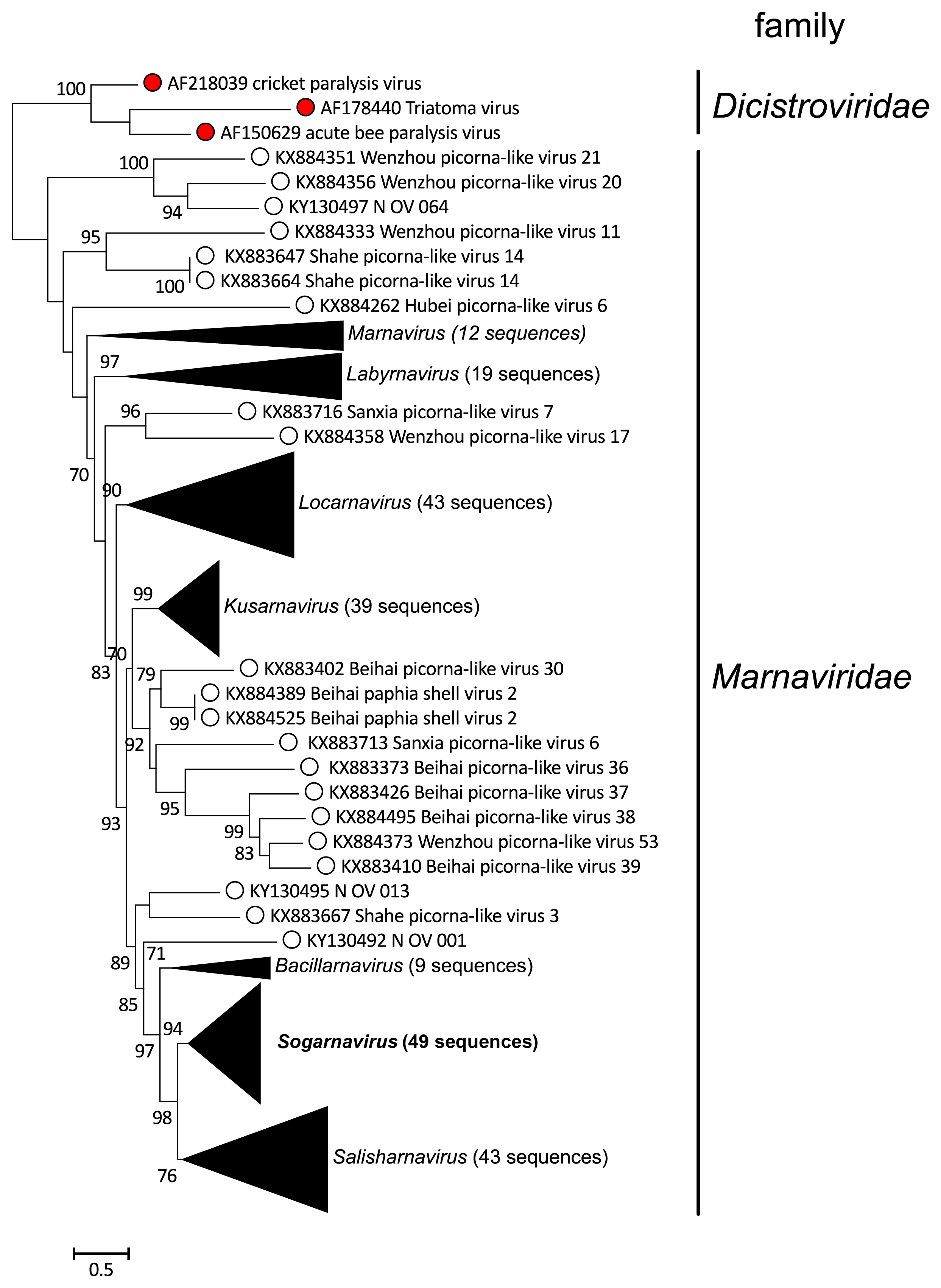 |
| Figure 5 Marnaviridae. Collapsed version of Figure 4. Marnaviridae. Collapsed genus clades include viruses classified into species in a genus and unclassified viruses also likely belonging to that genus. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Members of the family Marnaviridae share many properties with other viruses in the order Picornavirales. For example, their genomes are composed of one molecule of positive-sense RNA, have a 2C-3Cpro-3Dpol non-structural gene organization (Figure 2 Marnaviridae), and the virions of isolated representatives are icosahedral with a diameter of 22–35 nm (Figure 1 Marnaviridae; Table 2 Marnaviridae). However, phylogenetic analysis of RdRP amino acid sequences shows that members of the family Marnaviridae form a monophyletic group relative to viruses from other families within the Picornavirales (Figure 3 Marnaviridae).
Related, unclassified viruses
| Virus name | Accession number |
| Beihai paphia shell virus 1 strain BHZY60696 | KX884512 |
| Beihai paphia shell virus 1 strain BWBFG40339 | KX884527 |
| Beihai paphia shell virus 2 strain YYSZX17625 | KX884389 |
| Beihai paphia shell virus 2 strain BWBFG38484 | KX884525 |
| Beihai picorna-like virus 20 | KX883365 |
| Beihai picorna-like virus 30 | KX883402 |
| Beihai picorna-like virus 36 | KX883373 |
| Beihai picorna-like virus 37 | KX883426 |
| Beihai picorna-like virus 38 | KX884495 |
| Beihai picorna-like virus 39 | KX883410 |
| Hubei picorna-like virus 6 | KX884262 |
| N OV 001 | KY130492 |
| N OV 013 | KY130495 |
| N OV 064 | KY130497 |
| Sanxia picorna-like virus 6 | KX883713 |
| Sanxia picorna-like virus 7 | KX883716 |
| Shahe picorna-like virus 3 | KX883667 |
| Shahe picorna-like virus 14 strain SHWC0209c12767 | KX883647 |
| Shahe picorna-like virus 14 strain SHWC13617 | KX883664 |
| Wenling picorna-like virus 1 | KX884311 |
| Wenzhou gastropodes virus 1 | KX884372 |
| Wenzhou picorna-like virus 5 strain WZSLuoI84329 | KX884369 |
| Wenzhou picorna-like virus 5 strain BHZC35527 | KX884476 |
| Wenzhou picorna-like virus 11 | KX884333 |
| Wenzhou picorna-like virus 17 | KX884358 |
| Wenzhou picorna-like virus 20 | KX884356 |
| Wenzhou picorna-like virus 21 | KX884351 |
| Wenzhou picorna-like virus 53 | KX884373 |
Virus names and virus abbreviations are not official ICTV designations.
Metagenomic approaches targeting marine RNA viruses (Lachnit et al., 2015, López-Bueno et al., 2015, Moniruzzaman et al., 2017, Shi et al., 2016) have uncovered a diverse collection of viruses that are closely related to members of various families in the Picornavirales, including some that form a well-supported clade with members of the family Marnaviridae (Vlok et al., 2019). These are likely to represent additional members of the established genera as well as possible new genera (Figure 4 Marnaviridae, Figure 5 Marnaviridae).

