Family: Ovaliviridae
Li Huang and Haina Wang
The citation for this ICTV Report chapter is the summary published as Huang et al. (2021)
ICTV Virus Taxonomy Profile: Ovaliviridae, Journal of General Virology, 102 (3): 001546
Corresponding author: Li Huang (huangl@sun.im.ac.cn)
Edited by: Mart Krupovic and Stuart G. Siddell
Posted: November 2020
PDF: ICTV_Ovaliviridae.pdf
Summary
The family Ovaliviridae includes viruses with a linear dsDNA genome that replicate in hyperthermophilic archaea from the genus Sulfolobus (Table 1 Ovaliviridae). The virions of Sulfolobus ellipsoid virus 1 (SEV1) contain a protein capsid with 16 regularly spaced striations and are enveloped with a lipid membrane. Viral DNA, probably in the form of a nucleoprotein filament, wraps around the longitudinal axis of the virion in a plane to form a multilayered disk-like structure with a central hole, and sixteen of these structures are stacked to generate a spool-like capsid. Six-fold symmetrical virus-associated pyramids appear on the surface of SEV1-infected cells, which are ruptured to form a hexagonal opening for subsequent release of progeny virus particles. The unique shape and architecture of ovalivirus particles have not been observed among bacterial or eukaryotic viruses and are specific to archaeal viruses.
Table 1 Ovaliviridae. Characteristics of members of the family Ovaliviridae.
| Characteristic | Description |
| Example | Sulfolobus ellipsoid virus 1 (MF144115), species Alphaovalivirus fumarolicaense |
| Virion | Ellipsoid; 115 nm long, 78 nm wide; an envelope encases a striated capsid |
| Genome | Linear, dsDNA (23,219 bp) with 172 bp inverted terminal repeats |
| Replication | By a virus-encoded protein-primed family B DNA polymerase |
| Translation | Not characterized |
| Host range | Hyperthermophilic archaea of the genus Sulfolobus; lytic |
| Taxonomy | One genus with a single species |
Virion
Morphology
The SEV1 virion is ellipsoidal, measuring about 115 nm × 78 nm, and coated with an envelope (Figure 1 Ovaliviridae). As observed under cryo-EM, the SEV1 virion contains a striated capsid enclosed by an 11 nm-thick envelope decorated with protruding spikes (Figure 1B Ovaliviridae). Sixteen helical striations aligned perpendicular to the longitudinal axis of the particle with a periodicity of about 5 nm are clearly visible on the capsid. Each electron-dense stripe is about 2.8 nm wide. The longitudinal section of the 3-D cryo-ET of a virion reveals a tube-like structure of about 8 nm in diameter at the center of the capsid (Figure 1C Ovaliviridae and 1D Ovaliviridae). As shown in 2-D sectioned slices from the cryo-ET of the capsid, apparent striations are also evident in the interior of the particle. The capsid is probably formed by coiling of a nucleoprotein filament. The filament wraps tightly around the longitudinal axis multiple times in the middle portion of the capsid, in a plane, to form a disk-like structure (Figure 1C Ovaliviridae). Sixteen of the stacked disks constitute the SEV1 capsid (Figure 1D Ovaliviridae). The capsid is somewhat flexible in shape, presumably contributing to the constriction of the virion in the middle when the virus particle is dehydrated during analysis by negative-stain transmission electron microscopy (TEM; Figure 1A Ovaliviridae).
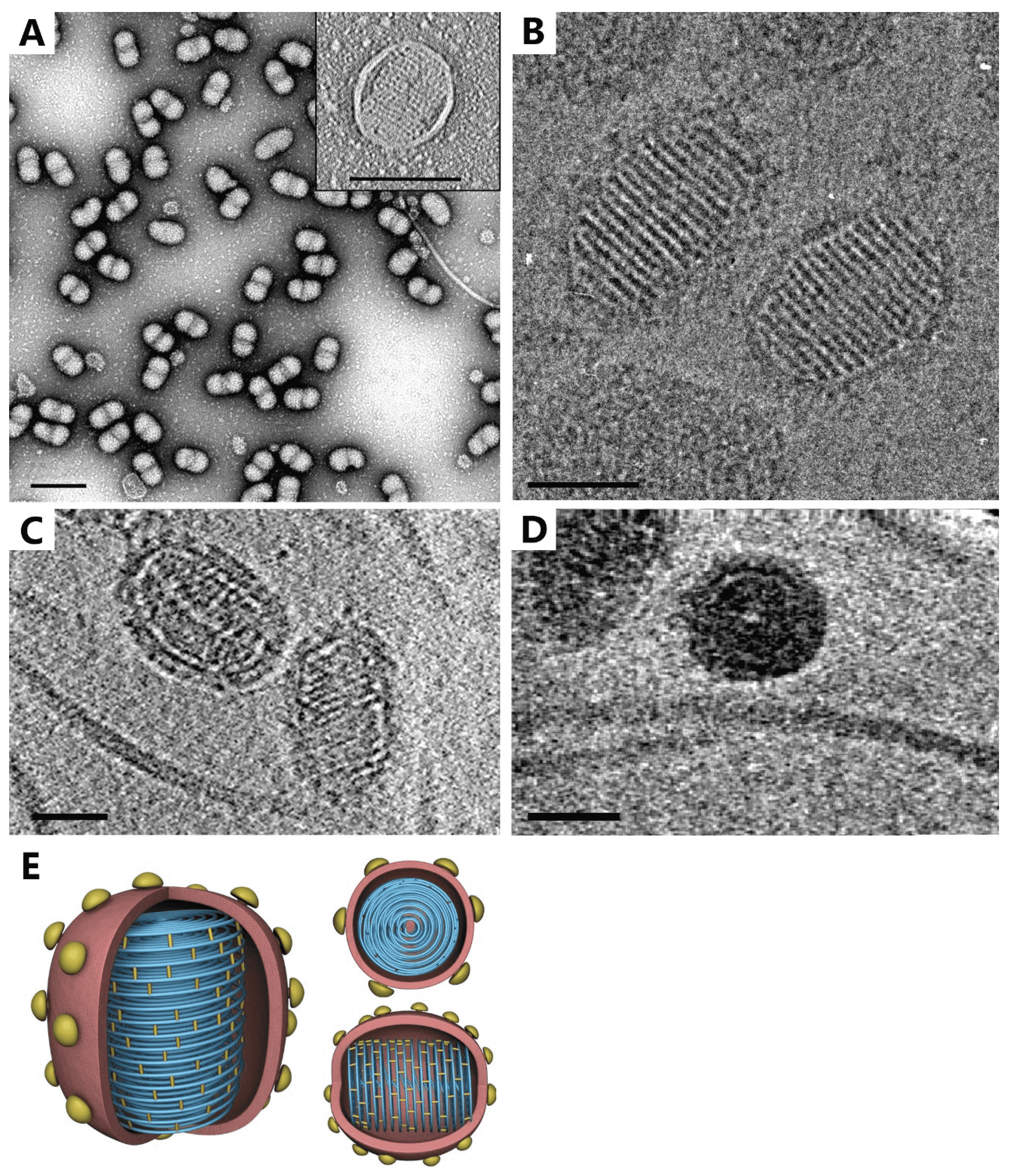 |
| Figure 1 Ovaliviridae. Virion structure of Sulfolobus ellipsoid virus 1 (A) Negative-staining electron micrographs of virions (Scale bar: 200 nm). Inset, a virion with capsid striations and an envelope (Scale bar: 100 nm). (B) Cryo-electron micrograph of virions (Scale bar: 50 nm). (C) A thin slice from the cryo-electron tomograph (Scale bar: 50 nm). (D) Top view (Scale bar: 50 nm). (E) Schematic 3-D model (modified from (Wang et al., 2018)). |
Physicochemical and physical properties
Virions are fragile and partially disassembled after high-speed ultracentrifugation. Virions are sensitive to organic solvents and detergents.
Nucleic acid
SEV1 virions contain a single molecule of double-stranded (ds) DNA of 23,219 bp with 172 bp inverted terminal repeats. The GC content of the genome is 33%.
Lipids
The SEV1 virion is enveloped, and the distribution pattern of isoprenoid glycerol dibiphytanyl glycerol tetraethers in the virion envelope is similar to that in the host cell membrane.
Proteins
SEV1 virions carry four major proteins of 12–170 kDa (Wang et al., 2018).
Genome organization and replication
The linear dsDNA genome of SEV1 is predicted to encode 38 proteins (Figure 2 Ovaliviridae) (Wang et al., 2018). The majority (27/38) of the open reading frames (ORFs) reside on one of the strands. Most ORFs encode proteins of unknown function. Four genes - vp1, vp2, vp3 and vp4 - encode the structural proteins of the virus. The genome also encodes a glycosyltransferase, a B-family DNA polymerase and a SAM-dependent methyltransferase. Four ORFs (the ORF for the SAM-dependent methyltransferase, ORF381, ORF100 and ORF140) are homologous to sequences in lipothrixviruses and/or rudiviruses, and the first three of them also to sequences in the spindle-shaped virus Sulfolobus monocaudavirus 1 (SMV1), an unclassified virus in the family Bicaudaviridae, implying possible horizontal gene transfer between these viruses. The putative B-family DNA polymerase, belonging to the subfamily of protein-primed DNA polymerases (Salas 1991), is similar to those from some plasmids and viruses including Acidianus bottle shaped virus (Ampullaviridae) (Peng et al., 2007), His1 virus (Halspiviridae) and His2 virus (Pleolipoviridae) (Bath et al., 2006), and Nitrosopumilus spindle-shaped virus 1 (Thaspiviridae) (Kim et al., 2019). This DNA polymerase is presumably responsible for the replication of the virus genome. However, no terminal protein, which is required to prime replication (Salas 1991), has been identified in Sulfolobus ellipsoid virus 1.
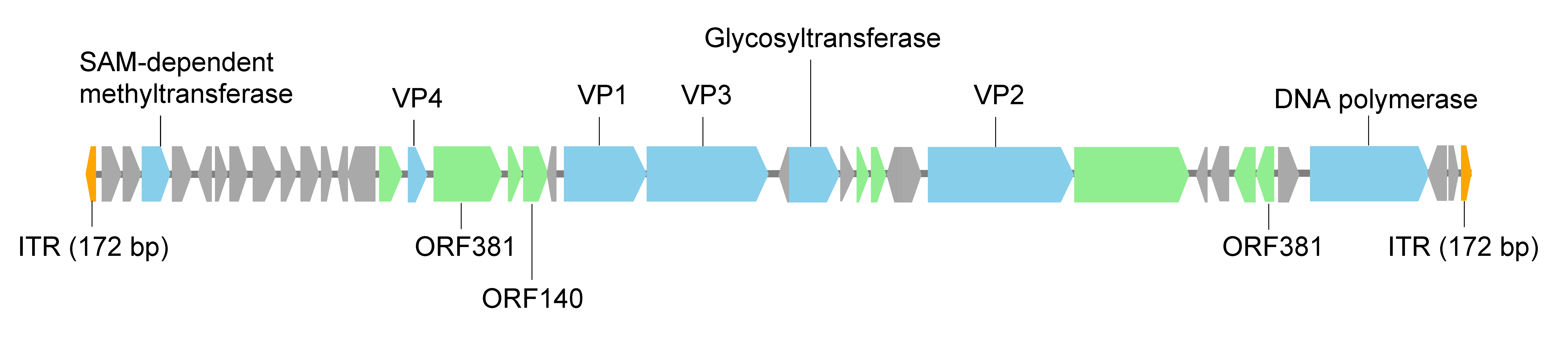 |
| Figure 2 Ovaliviridae. Sulfolobus ellipsoid virus 1 genome organization. The size and direction of each predicted ORF or genetic element is indicated by an arrow. Colours indicate ORFs with a putative function (blue), those homologous to ORFs from other archaeal viruses or containing conserved motifs (green), and those having no significant sequence matches to known sequences in public databases (grey). The inverted terminal repeats (ITRs) are coloured orange. |
Biology
SEV1 was isolated from an acidic hot spring (86−106oC, pH 2.2−2.5) in Laguna Fumarolica, Costa Rica. The only known host for SEV1 is Sulfolobus sp. A20 (Dai et al., 2016). Virus infection only slightly retards the growth of the host cells and yields no detectable cell debris. SEV1 virions are released through hexagonal openings on the host cell surface (Wang et al., 2018).
Derivation of names
Ovaliviridae: From Latin ovalis, for “oval”.
Sulfolobus ellipsoid virus 1: genus of archaeal host Sulfolobus and shape of virion.
Relationships within the family
No related viruses are known.
Relationships with other taxa
Most ovalivirus proteins have no known homologues. However, seven SEV1 ORFs, including one encoding a protein-primed DNA polymerase, show significant sequence matches to other archaeal viruses. Maximum likelihood phylogenetic analysis of the protein-primed DNA polymerases confirms that SEV1 is not closely related to any other archaeal or bacterial virus (Mart Krupovic, unpublished results).

