Family: Retroviridae
John Coffin, Jonas Blomberg , Hung Fan, Robert Gifford, Theodora Hatziioannou, Dirk Lindemann, Jens Mayer, Jonathan Stoye, Michael Tristem, Welkin Johnson
The citation for this ICTV Report chapter is the summary published as Coffin et al., (2021):
ICTV Virus Taxonomy Profile: Retroviridae 2021, Journal of General Virology 2021;102:001712
Corresponding author: Welkin Johnson ([email protected])
Edited by: Balázs Harrach and Peter Simmonds
Posted: November 2021
Summary
The family Retroviridae comprises two subfamilies and eleven genera. Retroviruses are widespread in nature and are found in a wide variety of vertebrate hosts. Virions are 80-100 nm in diameter, have a lipid envelope that displays the viral glycoproteins, and an inner protein core that contains the viral genome and replicative enzymes. The morphology of the inner core varies, and is often characteristic for viruses within the same genus. Replication involves reverse-transcription, with the viral positive-sense RNA genome serving as a template for the synthesis of double-stranded DNA, followed by integration into host-cell chromosomal DNA to produce a provirus. Integration into germline tissues/cells can give rise to heritable proviruses known as endogenous retroviruses (ERVs), and most vertebrate genomes contain thousands of ERV loci.
Table 1. Retroviridae. Characteristics of members of the family Retroviridae
| Characteristic | Description |
| Example | Moloney murine leukemia virus (AF033811), species Gammaretrovirus murleu, subfamily Orthoretrovirinae |
| Virion | Enveloped spheres of 80–100 nm diameter with 8 nm glycoprotein spikes |
| Genome | Dimer of ssRNA of 7–13 kb (orthoretroviruses), or ssRNA (partially reverse-transcribed in the virion to dsDNA) of 11–12 kb (spumaretroviruses) |
| Replication | dsDNA produced by reverse transcription of the RNA genome is integrated into host genome and serves as template for synthesis of the virus genome by cellular RNA polymerase II |
| Translation | From capped and polyadenylated genomic transcripts and subgenomic, spliced mRNAs |
| Host range | Vertebrates |
| Taxonomy | Realm Riboviria, kingdom Pararnavirae, phylum Artverviricota, class Revtraviricetes, order Ortervirales; 2 subfamilies (Orthoretrovirinae and Spumaretrovirinae), 11 genera including 65 species |
Virion
Morphology
Virions are spherical, enveloped and 80–100 nm in diameter (Vogt 1997). Glycoprotein surface projections are about 8 nm in length, irregularly spaced. The internal capsid core comprises assembled capsid proteins encompassing a complex of nucleocapsid proteins and viral nucleic acid. The apparently spherical nucleocapsid (nucleoid) is eccentric for members of the genus Betaretrovirus, concentric for members of the genera Alpharetrovirus, Gammaretrovirus, Deltaretrovirus, and rod or truncated cone-shaped for members of the genus Lentivirus. Members of the subfamily Spumaretrovirinae have concentric, spherical nucleocapsids (Figure 1. Retroviridae).
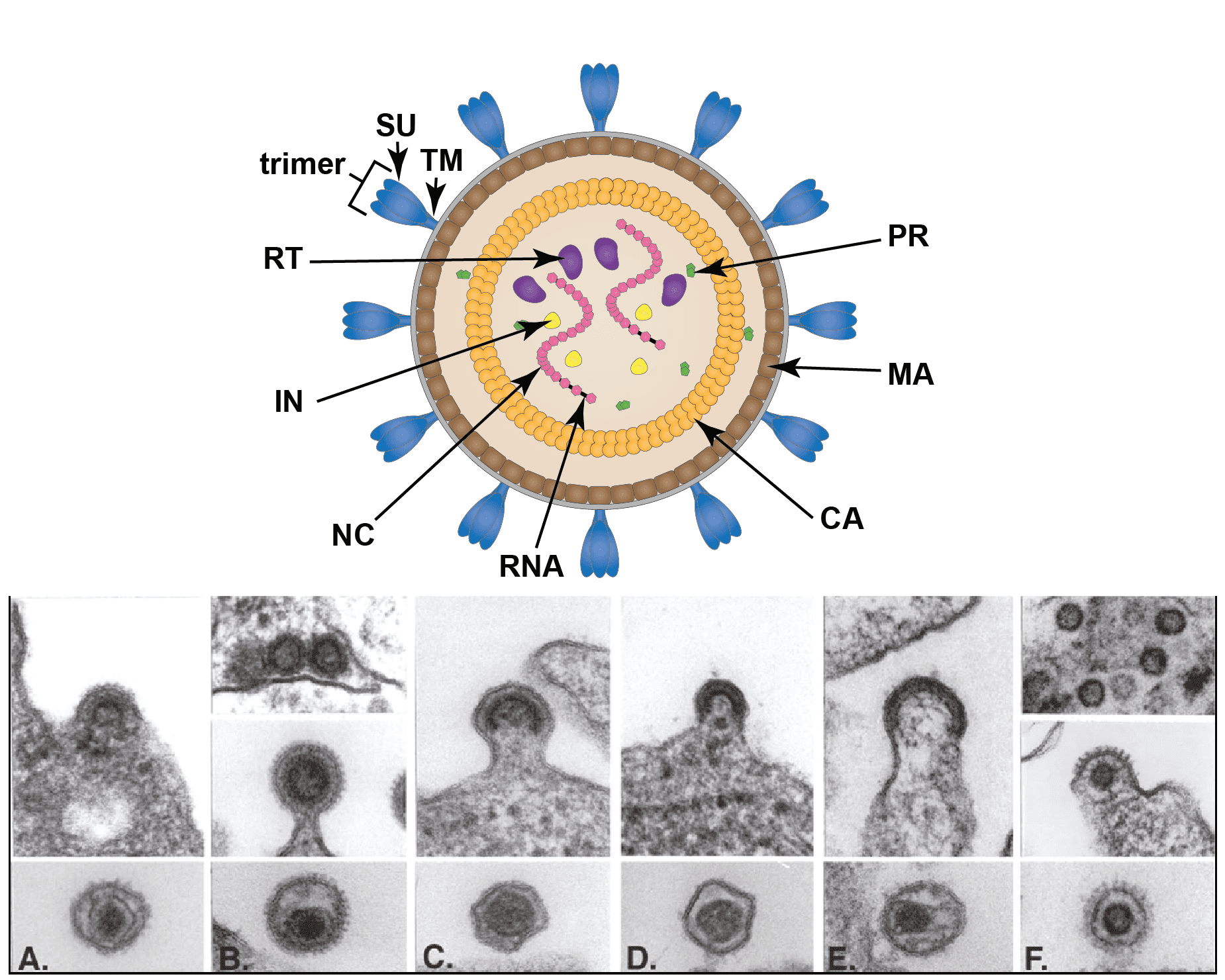 |
| Figure 1. Retroviridae. Structure of retrovirus particles. (Top) Schematic cartoon (not to scale) showing the inferred locations of various structures and proteins common to retroviral virions (courtesy of B. Lawhorn). MA – matrix; CA- capsid; NC – nucleocapsid; PR – protease; RT- reverse transcriptase; IN -integrase; SU – surface subunit; TM – transmembrane subunit. (Bottom) Budding and mature virus particles. Panel (A): avian leukosis virus (genus Alpharetrovirus); type “C” morphology. Panel (B): mouse mammary tumor virus (Betaretrovirus); type “B” morphology. Panel (C): murine leukemia virus (Gamamaretrovirus). Panel (D): bovine leukemia virus (Deltaretrovirus). Panel (E): human immunodeficiency virus 1 (Lentivirus). Panel (F): simian foamy virus Pan troglodytes schweinfurthii (SFVpsc; formerly called PFV or HFV) (Simiispumavirus , subfamily Spumaretrovirinae). (Courtesy of M. Gonda, reproduced with permission from (Coffin et al., 1997)). |
Two distinct morphogenic pathways exist. Historically, a nomenclature based on electron microscopy classified members of the Alpharetrovirus and Gammaretrovirus genera, which assemble their immature capsids at the plasma membrane, as C-type viruses. Members of the Betaretrovirus genus in contrast were said to assemble A-type particles (immature capsids) in the cytoplasm which then budded with either a B-type (mouse mammary tumor virus, MMTV) or D-type (Mason-Pfizer monkey virus, MPMV) morphology. This scheme is no longer used for classification, although the morphological descriptions still sometimes appear.
Physicochemical and physical properties
Virion buoyant density is 1.16–1.18 g cm−3 in sucrose density gradients. Virion S20,w is approximately 600S in sucrose density gradients. Virions are sensitive to heat, detergents and formaldehyde. The surface glycoproteins may be partially removed by proteolytic enzymes or in some cases by mechanical forces (e.g. vortexing, centrifugation). Virions are relatively resistant to UV light.
Nucleic acid
A typical virus genome of a member of the subfamily Orthoretrovirinae consists of a homodimer of linear, positive sense, ssRNA, each monomer of 7–13 kb (Goff 2013). The RNA constitutes about 2% of the virion dry weight. The monomers are held together by hydrogen bonds. Each monomer of RNA is polyadenylated at the 3′-end and has a 7-methyl cap structure (type 1) at the 5′-end. The purified virion RNA is not infectious. Each monomer is associated with a specific molecule of tRNA that is base-paired to a region (termed the primer binding site, PBS) near the 5′-end of the RNA and involves about 18 nt at the 3′-end of the tRNA. Other host-derived RNAs (and small DNA fragments) found in virions are believed to be incidental inclusions. The virions of members of the subfamily Spumaretrovirinae contain some dsDNA since reverse transcription of the packaged viral RNA genome occurs in 5–10% of released virus particles. The exact structure of the DNA has not been determined.
Proteins
Proteins constitute about 60% of virion dry weight. A standard nomenclature for retrovirus proteins and protein domains has been adopted (Leis et al., 1988). There are two envelope proteins: SU (surface) and TM (transmembrane) encoded by the viral env gene. Some members of the subfamily Spumaretrovirinae have a third Env protein, LP (leader peptide). There are 3–6 internal, non-glycosylated structural proteins (encoded by the gag gene). These include, in order from the amino terminus, (1) MA (matrix), (2) CA (capsid protein) and (3) NC (nucleocapsid). A fourth protein, thought to be involved in virion budding, varies in position within Gag. The MA protein is often acylated with a myristyl moiety covalently linked to the amino-terminal glycine. Other proteins are a protease (PR, encoded by the pro gene), a reverse transcriptase (RT, encoded by the pol gene) and an integrase (IN, encoded by the pol gene). In some viruses, a dUTPase (DU, role uncertain) is also present. Members of the Spumaretrovirinae encode only a single Gag protein which is cleaved once near the carboxyl-terminus in about half of the proteins. The virions of primate lentiviruses also specifically incorporate additional small, virally-encoded accessory proteins, such as p6, Vpr and Vpx, that function during the early, post-entry stage of target cell infection.
Lipids
Lipids constitute about 35% of virion dry weight and are derived from the plasma membrane of the host cell.
Carbohydrates
Virions are composed of about 3% carbohydrate by weight. This value varies, depending on the virus. At least one (SU), and usually both envelope proteins are glycosylated. In the case of spumaviruses, the Env leader peptide is also glycosylated. Cellular glycolipids and some glycoproteins are also found in the virion envelope.
Genome organization and replication
For all retroviruses, reverse transcription of an RNA genome produces a double-stranded DNA with long terminal repeats (LTRs) (Figure 2. Retroviridae) (Telesnitsky and Goff 1997). The dsDNA is then integrated into the host cell genome, resulting in formation of a DNA provirus (Figure 3. Retroviridae). The provirus serves as a template for synthesis of the viral genome and mRNAs by cellular RNA polymerase II (Goff 2013, Brown 1997). Virions of members of the subfamily Orthoretrovirinae carry two copies of the RNA genome. Infectious viruses have four main genes coding for the virion proteins in the order: 5′-gag-pro-pol-env-3′. Some retroviruses contain genes encoding non-structural proteins important for the regulation of gene expression and virus replication. Others carry cell-derived sequences that are important in carcinogenesis (oncogenes) (Rosenberg and Jolicoeur 1997). These cellular sequences are inserted either into a complete retrovirus genome (some strains of the alpharetrovirus Rous sarcoma virus) or in the form of substitutions for deleted viral sequences (all other transforming alpharetroviruses and gammaretroviruses). Such deletions render the virus replication-defective and dependent on non-transforming helper viruses for production of infectious progeny. In many cases the cell-derived sequences are expressed as a fused gene with a viral structural gene that is then translated into one chimeric protein (e.g. Gag-Onc protein).
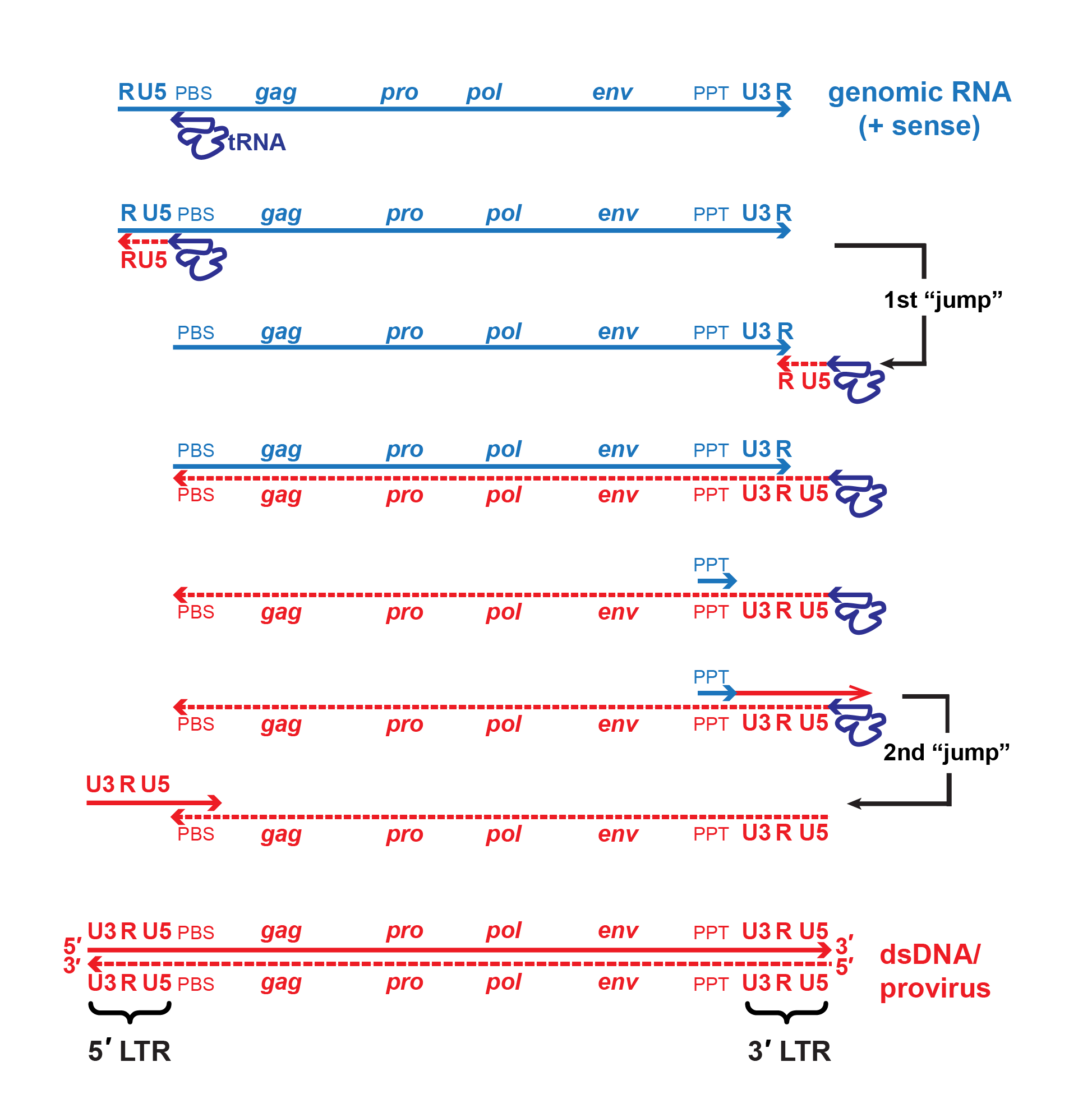 |
| Figure 2. Retroviridae. Mechanism of reverse transcription. A single copy of the positive-sense viral RNA genome is shown in blue, DNA intermediates and dsDNA are shown in red. The canonical viral RNA genome structure is: 5′-R-U5-PBS-gag-pro-pol-env-PPT-U3-R-3′. Reverse transcriptase initiates negative-strand DNA synthesis using a cellular tRNA primer complementary to the primer binding site (PBS). Synthesis proceeds towards the 5′-end of the RNA template, then the growing template “jumps” to the 3′-end of the same or another RNA genome (only one is shown for simplicity) mediated by complementarity of the R regions. During minus-strand synthesis, the RNA template is digested by the viral RNAse H, leaving a short, RNAse H-resistant fragment complementary to the polypurine tract (PPT) which serves as the primer for positive-strand DNA synthesis. Some viruses may also initiate positive-strand synthesis at a central polypurine tract, or CPPT. Note that the cellular tRNA primer also serves as the template for regenerating the PBS. A second template switch occurs when the elongated positive-strand jumps to the 3′-end of the minus-strand template via complementarity provided by the PBS region. Extension of both strands produces the final double-stranded DNA molecule with complete 5′- and 3′-LTRs (U3-R-U5). |
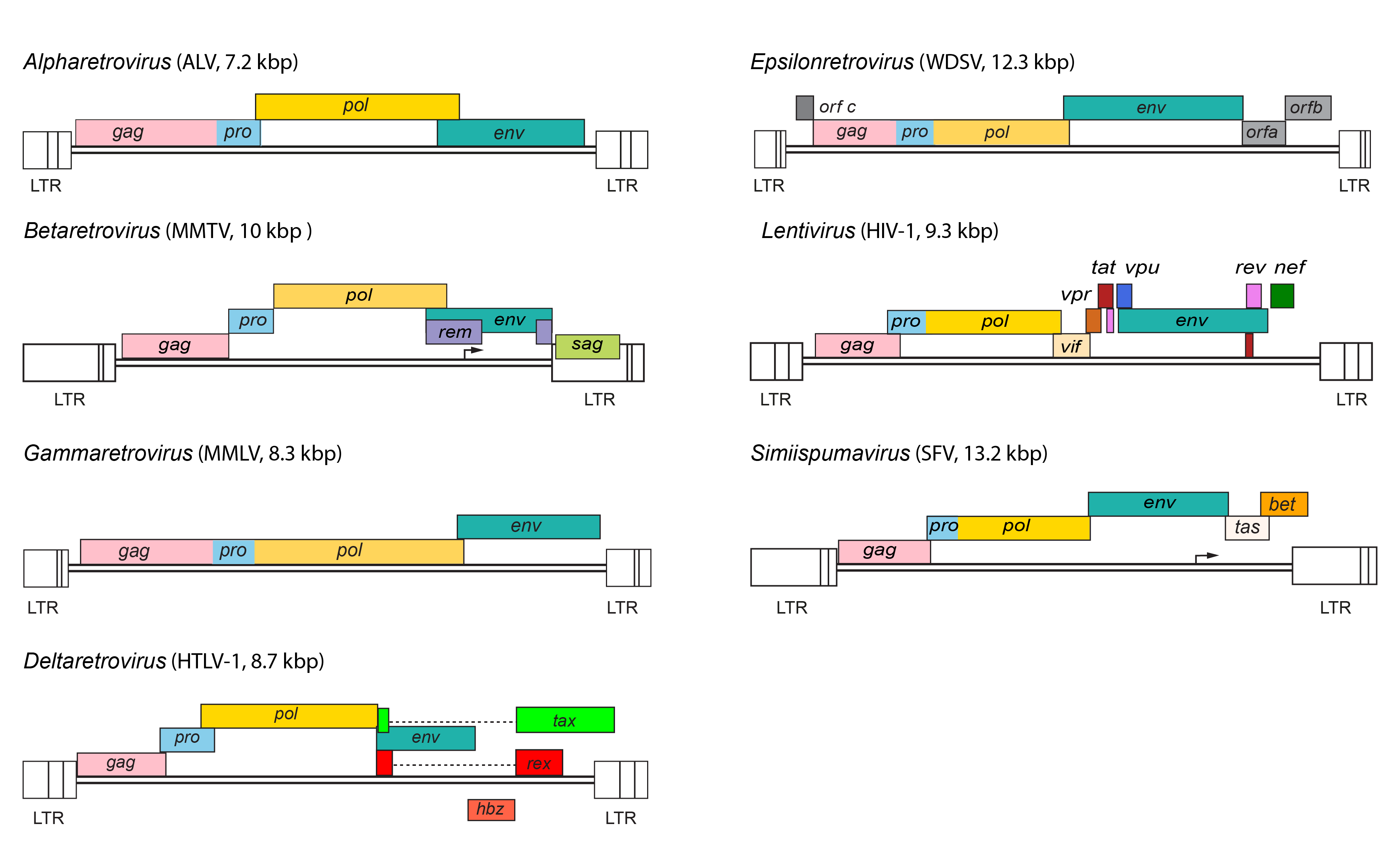 |
| Figure 3. Retroviridae. Provirus structures for representative viruses of the Alpharetrovirus, Betaretrovirus, Gammaretrovirus, Deltaretrovirus, Epsilonretrovirus, Lentivirus and Simiispumavirus genera. |
Entry into the host cell is mediated by interaction between the virion SU glycoprotein and specific receptors at the host cell surface, resulting in fusion of the viral envelope with the plasma membrane, either directly or following endocytosis. Retrovirus receptors are cell surface proteins (Greenwood et al., 2018, Overbaugh et al., 2001). For human immunodeficiency virus (HIV), both the CD4 protein, which is an immunoglobulin-like molecule with a single transmembrane region, and a chemokine receptor (CCR5 or CXCR4), which spans the membrane seven times, are required for membrane fusion. The receptors for gammaretroviruses are involved in the transport of small molecules and have a complex structure with multiple transmembrane domains; almost all gammaretrovirus receptors identified to date are classified as solute carrier (SLC) proteins (Greenwood et al., 2018). For the avian leukosis viruses (ALVs, genus Alpharetrovirus), four receptors have been identified: the receptor for subgroup A viruses is a small protein with a single transmembrane domain that is distantly related to a cell receptor for low-density lipoprotein, that for subgroup B viruses is related to the TNF-receptor family of proteins, that for subgroup C viruses is related to the mammalian butyrophilins, and that for subgroup J viruses is the chicken Na+/H+ exchanger protein (Barnard et al., 2006).
The process of intracellular uncoating of viral particles is not well understood. Early post-entry events are carried out in the context of a capsid core delivered to the cytoplasm.
For members of the subfamily Orthoretrovirinae, replication starts with reverse transcription (by RT) of virion RNA into cDNA using the 3′-end of the tRNA bound to the primer binding site as primer for synthesis of a negative-sense cDNA transcript (Figure 2. Retroviridae). The initial short product (to the 5′-end of the genome) transfers and primes further cDNA synthesis from the 3′-end of the genome by virtue of duplicated (R) sequences at the ends of the viral RNA. cDNA synthesis involves the concomitant digestion of the viral RNA (RNase H activity of the RT protein). A product of this hydrolysis (PPT) serves to prime positive-sense cDNA synthesis on the negative-sense DNA copies. The resulting short product ends in the tRNA primer, and is transferred to the end of the minus-strand cDNA by virtue of the tRNA duplication, thus priming completion of the plus-strand. In its final form, the linear dsDNA derived from the viral ssRNA genome contains long terminal repeats (LTRs) composed of unique sequences from the 3′- (U3) and 5′- (U5) ends of the viral RNA flanking the R sequence. Reverse transcription is thought to follow the same pathway in members of the subfamily Spumaretrovirinae, but the timing is different as it occurs during viral assembly or release from the cell (Rethwilm 2010).
The process of reverse transcription is characterized by a high frequency of recombination due to the transfer of the RT from one template RNA to the other. The mechanism of reverse transcription and lack of proofreading allow for high rates of recombination and genetic diversity for many of the retroviruses. The high rate of genetic variation in vivo can lead to formation of a complex population or “swarm” consisting of a large number of genetically diverse viruses.
Retroviral DNA becomes integrated into the chromosomal DNA of the host, to form a provirus, by a mechanism involving the viral IN protein. The ends of the virus DNA are joined to host DNA, involving the removal of two bases from the ends of the linear viral DNA and generating a short duplication of the host sequence at the integration site. Virus DNA can integrate at many sites in the host genome. However, once integrated, a provirus is incapable of further autonomous transposition within the same cell. The genome of the integrated provirus is co-linear with that of non-integrated viral DNA, with the addition of U3 and U5 sequences at the 5′- and 3′- ends respectively. Integration appears to be a prerequisite for virus replication. Different retroviruses can show distinct preferences in integration site selection; for example, human immunodeficiency virus 1 (HIV-1) tends to insert within actively transcribed gene sequences whereas members of the species Murine leukemia virus preferentially integrate in regions near the start of transcribed genes (Kvaratskhelia et al., 2014).
The integrated provirus is transcribed by cellular RNA polymerase II into virion RNA and mRNA species in response to transcriptional signals in the viral LTRs. The genomes of complex retroviruses in the Betaretrovirus, Deltaretrovirus, Epsilonretrovirus, Lentivirus genera and in the Spumavirinae subfamily also encode non-structural proteins, including transcriptional transactivators, which are required for expression of the LTR promoters, as well as proteins required for efficient export of unspliced viral RNA from the nucleus. Additional nonstructural proteins expressed by members of some genera are also involved in protection against host defenses.
There are several classes of mRNA depending on the virus and its genetic map. An mRNA comprising the whole genome serves for translation of the gag, pro and pol genes (positioned in the 5′-half of the RNA). This step results in the formation of two polyprotein precursors, Gag and Gag-Pro-Pol, which are cleaved to yield the structural proteins (from Gag) as well as the PR, RT and IN enzymes (from Pro-Pol). A smaller mRNA consisting of sequences corresponding to the 5′-end of the genome spliced to sequences from the 3′-end of the genome, and including the env gene and the U3 and R regions, is translated into the precursor of the envelope proteins. In viruses that contain additional genes, other forms of spliced mRNA are also made; all these spliced mRNAs share a common leader sequence at their 5′ ends. Members of the subfamily Spumaretrovirinae are unique in that they make use of an internal promoter (IP), which is located in the env gene for transcription of distal accessory genes (Figure 4. Retroviridae). Most primary translation products in retrovirus infections are polyproteins that require proteolytic cleavage before becoming functional. The gag, pro and pol gene products are generally produced from a nested set of primary translation products. For pro and pol, translation involves bypassing translational termination signals either by ribosomal frameshifting (e.g. HIV-1) or by readthrough of stop codons at the Gag-Pro and/or the Pro-Pol boundaries (e.g. members of the species Murine leukemia virus). Members of the subfamily Spumaretrovirinae synthesize Pro and Pol proteins from their own spliced mRNA rather than as a Gag-Pro-Pol fusion protein (Figure 4. Retroviridae).
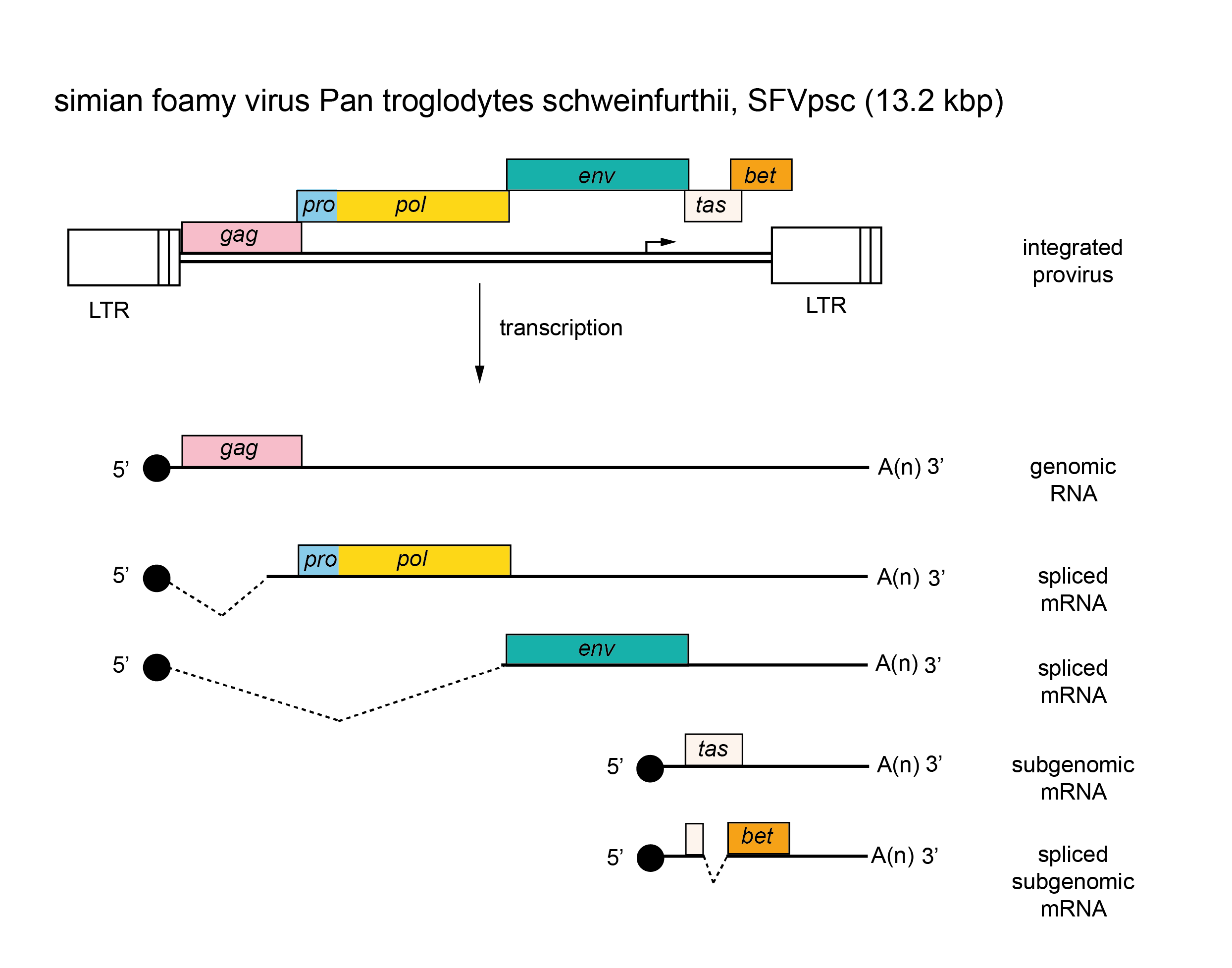 |
| Figure 4. Retroviridae. Spumaretrovirus genome organization. The 13.2 kbp simian foamy virus Pan troglodytes schweinfurthii (SFVpsc) provirus is shown with LTRs, the internal promoter, protein-coding regions (gag, pro, pol, env, tas and bet) and transcripts (solid line arrows with protein names added) marked. |
The retroviral genomic RNA contains sequences of varying lengths, usually located in the 5′-end leader U3 and in gag, which include a packaging signal (Ψ) (Johnson and Telesnitsky 2010). Ψ is required for efficient encapsidation of the genome into particles, and is generally not present on the subgenomic mRNAs, a notable exception being the alpharetroviruses. In the case of the spumaviruses, Ψ does not appear to be in the 5′-end of the genome. In all well-studied cases, Ψ activity is not defined by the primary sequence, but by a complex, folded RNA structure.
Capsids assemble either at the plasma membrane (for a majority of genera), or as intracytoplasmic particles (for members of the subfamily Spumaretrovirinae and members of the genus Betaretrovirus). Virions are released from the cell by the processes of budding and maturation (Pornillos and Ganser-Pornillos 2019). Budding appears to occur preferentially at specialized membrane microdomains known as lipid rafts. Virions of the spumaviruses and deltaretroviruses are highly cell-associated. Processing of the polyprotein precursor to the internal proteins occurs concomitant with or just subsequent to the maturation of virions of members of the subfamily Orthoretrovirinae.
Biology
Retroviruses are widely distributed as exogenous infectious agents of vertebrates (Goff 2013). In addition, endogenous proviruses that have resulted from infection of germ line cells are inherited as Mendelian genes, and are commonly referred to as endogenous retroviruses (ERVs) (Johnson 2019, Stoye 2012). ERV loci occur widely among vertebrates and can constitute up to 10% of genomic DNA. ERV sequences can be readily incorporated into retrovirus phylogenies, and frequently cluster with exogenous retroviruses within existing genera – greatly enhancing reconstruction of the natural history and origins of the Retroviridae lineage (Gifford 2012, Jern et al., 2005). The presence of ERV loci in most or all vertebrate genomes indicates that their current very broad distribution among vertebrate hosts extends back hundreds of millions of years (Aiewsakun and Katzourakis 2015, Aiewsakun and Katzourakis 2017). The vast majority of ERVs have suffered inactivating mutations and cannot produce infectious virus. A few can exert significant biological effects following activation, either by replication in a manner indistinguishable from exogenous viruses or following recombination with replication-competent virus. There are also numerous examples of ERV loci that have been coopted for cellular/host functions; examples include exapted viral proteins (e.g. synctyins) and LTRs serving as cis-acting regulatory elements (Chuong et al., 2017, Lavialle et al., 2013).
Retroviruses are associated with a variety of diseases (Rosenberg and Jolicoeur 1997, Maeda et al., 2008). These include: malignancies, including certain leukemias, lymphomas, sarcomas and other tumors of mesodermal origin; mammary carcinomas and carcinomas of liver, lung and kidney; immunodeficiencies (such as AIDS); autoimmune diseases; lower motor neuron diseases; and several acute diseases involving tissue damage. Some retroviruses appear to be non-pathogenic. Transmission of retroviruses is horizontal via a number of routes, including blood, saliva, sexual contact, etc., and via direct infection of the developing embryo, or via milk or perinatal routes. Endogenous retroviruses are transmitted vertically by inheritance of germline proviruses.
Antigenicity
Virion proteins contain type-specific and group-specific determinants. Some type-specific determinants of the envelope glycoproteins are involved in antibody-mediated virus neutralization. Group-specific determinants are shared by members of a serogroup and may be shared between members of different serogroups within a particular genus. There is evidence for weak cross-reactivities between members of different genera. Epitopes that elicit T-cell responses are found on many of the structural proteins. Antigenic properties are not used in classification of members of the family Retroviridae.
Subfamily demarcation criteria
The two subfamilies are defined primarily based on comparison of amino-acid sequences of conserved regions of the Pol polyprotein, particularly the RT protein (Jern et al., 2005, Xiong and Eickbush 1990), as well as some differences in gene expression (e.g. Pol is expressed from a separate transcript for members of the Spumaretrovirinae and as part of the Gag-Pro-Pol polyprotein for members of the Orthoretrovirinae), presence/absence of subdomains in the Gag protein, and the timing of reverse-transcription relative to the replication cycle.
Genus demarcation criteria
Phylogenetic trees based on conserved domains of reverse transcriptase (RT) are conveniently rooted using RT from viruses and retroelements outside the family Retroviridae, and can be used to resolve relationships between genera in both subfamilies. Genera can also be distinguished based on the presence or absence of specific regulatory or accessory proteins, unique features of the Env protein, and comparison of amino-acid sequences of the TM subunit of Env. Parallel comparison of RT and TM can also be used to identify recombination events between viruses of different genera; for example, some viruses classified as betaretroviruses encode an Env protein typically associated with gammaretroviruses. Trees depicting relationships between members of different genera in the subfamily Spumaretrovirinae often mirror host phylogeny (indicative of stable, long-term virus-host relationships), and classification into genera is based in part on host association. Incorporation of sequences derived from vertebrate genomic ERV loci into RT-based phylogenies indicate that there may be extant (or possibly extinct) exogenous retroviruses that would form the basis for creation of additional genera (Gifford et al., 2018, Hayward et al., 2015).
Derivation of names
Alpharetrovirus: from a (alpha), first letter of the Greek alphabet.
Betaretrovirus: from b (beta), second letter of the Greek alphabet.
Bovispumavirus: from the association of these spumaviruses with bovine hosts.
Deltaretrovirus: from d (delta), fourth letter of the Greek alphabet.
Epsilonretrovirus: from e (epsilon), fifth letter of the Greek alphabet.
Equispumvirus: from the association of these spumaviruses with equine hosts.
Felispumavirus: from the association of these spumaviruses with feline hosts.
Gammaretrovirus: from g (gamma), third letter of the Greek alphabet.
Lentivirus: from Latin lentus, “slow”.
Orthoretrovirinae: from Greek orthos, “straight”.
Prosimiispumavirus: from the association of these spumaviruses with prosimian hosts.
Retrovirdae: from Latin retro, “backwards”, refers to the activity of reverse transcriptase which transfers genetic information from RNA “back” to DNA.
Simiispumavirus: from the association of these spumaviruses with simian hosts.
Spumaretrovirinae: from Latin spuma, “foam”, in reference to a characteristic “foamy” cytopathic effect displayed by infected cells in culture.
Relationships within the family
The division into two subfamilies is strongly supported by phylogenetic analysis based on highly conserved domains of reverse-transcriptase (Figure 5. Retroviridae). All viruses currently assigned to the subfamily Spumaretrovirinae share several common features that are absent from viruses of the subfamily Orthoretrovirinae (see discussion under subfamily description). Notably, phylogenetic analyses that include endogenous retrovirus (ERV) sequences found in the genomes of a wide range of vertebrates also support the subfamily division.
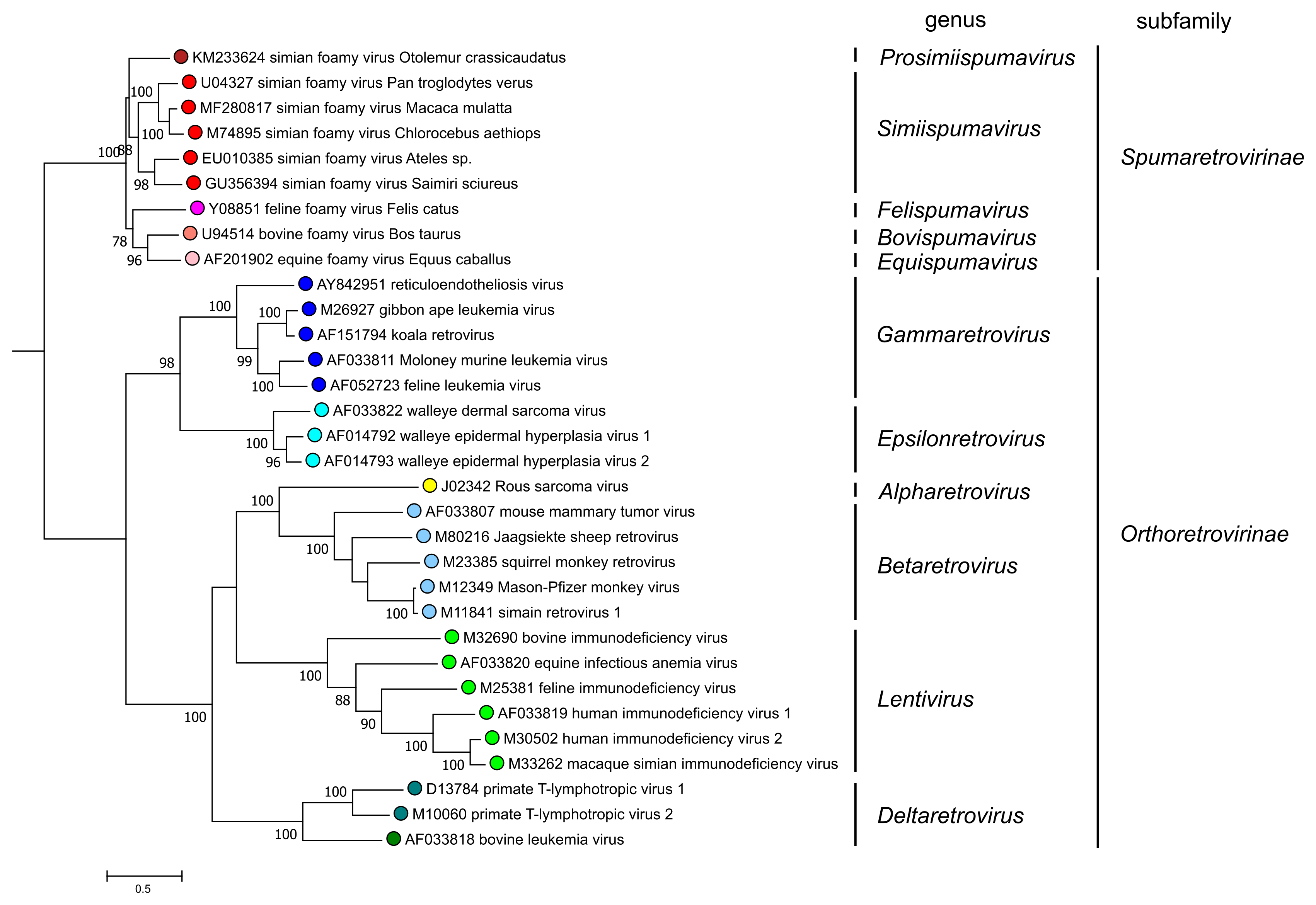 |
| Figure 5. Retroviridae. Phylogenetic relationships of selected retroviruses based on an amino-acid alignment spanning reverse transcriptase and the NTD and CCD domains of integrase (Xiong and Eickbush 1990, Lesbats et al., 2016). An unrooted phylogenetic tree was generated by maximum likelihood (PhyML3.2.2) (Guindon et al., 2010, Guindon and Gascuel 2003); this tree was subsequently rooted for clarity at the node-separating viruses of the Orthoretrovirinae and Spumaretrovirinae subfamilies. Numbers next to nodes indicate bootstrap support (100 replicates). Colored circles correspond to genera within each subfamily (5 in the Spumaretrovirinae and 6 in the Orthoretrovirinae). This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
Deep phylogenetic relationships to members of the other families in the Ortervirales are apparent for the reverse transcriptase (RT) protein (Xiong and Eickbush 1990, Krupovic et al., 2018). Viruses of the Caulimoviridae, Metaviridae, Pseudoviridae and Belpaoviridae families may also share other characteristics with retroviruses, including the presence of capsid (CA) and nucleocapsid (NC) domains, a protease (PR), and the use of a cellular tRNA to prime viral genome replication (Krupovic et al., 2018). Similar to retroviruses, members of the families Metaviridae, Pseudoviridae and Belpaoviridae encode an integrase (IN) and possess long-terminal repeats (LTRs).

