Family: Solemoviridae
Merike Sõmera, Denis Fargette, Eugénie Hébrard and Cecilia Sarmiento
The citation for this ICTV Report chapter is the summary published as Sõmera et al,. 2021
ICTV Virus Taxonomy Profile: Solemoviridae 2021, Journal of General Virology 2021;102:001707
Corresponding author: Merike Sõmera (merike.somera@taltech.ee)
Edited by: F. Murilo Zerbini, Sead Sabanadzovic and Luisa Rubino
Posted: January 2021, updated November 2021
PDF: ICTV_Solemoviridae.pdf (January 2021 version)
Summary
Plant viruses in the family Solemoviridae have stable icosahedral particles (20–34 nm in diameter) assembled on T=3 symmetry and a relatively small (4–6 kb) positive-sense, monopartite polycistronic RNA genome with a virus protein-genome linked (VPg) covalently attached to the 5′-terminus and no poly(A) tract at the 3′-terminus (Table 1). Replication is presumably cytoplasmic. Several solemoviruses support replication and encapsidation of small circular satellite RNAs. Solemovirus genomes have 4–10 open reading frames (ORFs). These ORFs code for a viral suppressor of RNA silencing, a polyprotein and a capsid protein. Domains of the polyprotein comprise a membrane anchor, a serine protease, a VPg and either a C-terminal domain protein or an RNA-directed RNA polymerase (RdRP; expressed by means of ribosomal frameshifting). These two variants of the polyprotein are translated via a ribosomal leaky scanning mechanism from genomic RNA and they undergo proteolytic processing at conserved cleavage sites between the domains. The capsid protein (CP) is translated from a subgenomic RNA. The natural host range is relatively narrow. Transmission occurs via mechanical wounding, by vegetative propagation or abiotically through soil or by insects (beetles, aphids, thrips, hoppers, mirid bugs and moths). Poleroviruses and enamoviruses are almost exclusively transmitted by aphids in persistent, circulative and non-propagative manner. Viruses infecting legumes or plants from the family Chenopodiaceae may be seed-transmissible. Members of the family are classified into four genera: Sobemovirus, Polemovirus, Polerovirus and Enamovirus.
Table 1. Solemoviridae. Characteristics of members of the family Solemoviridae
| Characteristic | Description |
| Example: | southern bean mosaic virus (DQ875594), species Sobemovirus SBMV |
| Virion | Non-enveloped icosahedral particles with T=3 symmetry, 20–34 nm in diameter, comprised of 180 molecules of capsid protein |
| Genome | 4–6 kb positive–sense, non-segmented RNA, with 5′-terminal VPg, no poly(A) tail |
| Replication | Cytoplasmic |
| Translation | Directly from genomic RNA by leaky scanning, ribosome entry site mediated initiation, −1 ribosomal frameshifting, polyprotein proteolytic processing; 3′ co-terminal subgenomic mRNA(s) are translated via leaky scanning and readthrough of an amber stop codon |
| Host range | Plants (monocotyledons and dicotyledons) |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Pisuviricota, class Pisoniviricetes, order Sobelivirales; five genera including 120 species |
Virion
Morphology
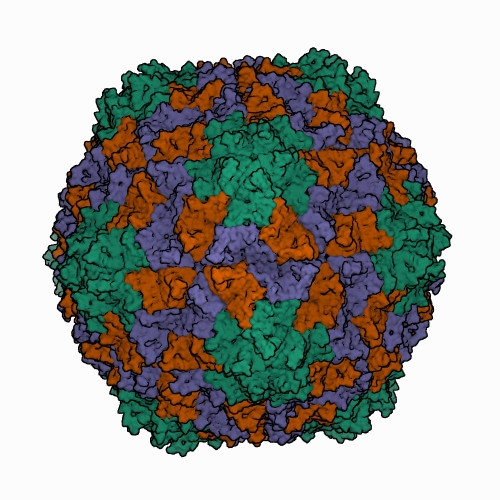
Icosahedral virions of sobemoviruses are 26–32 nm in diameter assembled on a T=3 lattice symmetry, without a lipid envelope. Detailed virion structures have been determined for several sobemoviruses: southern cowpea mosaic virus (SCPMV, initially recognized as the southern bean mosaic virus C-strain) at 2.8 Å resolution (Abad-Zapatero et al., 1980), southern bean mosaic virus (SBMV, initially recognized as the SBMV B-strain) at 2.9 Å resolution (Silva and Rossmann 1987), Sesbania mosaic virus (SeMV) at 3 Å resolution (Silva and Rossmann 1987), rice yellow mottle virus (RYMV) at 2.8 Å resolution (Qu et al., 2000), cocksfoot mottle virus (CfMV) at 2.7 Å (Tars et al., 2003), and ryegrass mosaic virus (RGMoV) at 2.9 Å resolution (Plevka et al., 2007). The 3D structures of different sobemovirus virions are nearly identical - their superimposed backbone C-α atoms show root mean square deviations (RMSDs) of 1.4–1.9 Å (Plevka et al., 2007).The virion of the only known polemovirus poinsettia latent virus (PnLV) is 34 nm in diameter, being slightly larger than that of sobemoviruses. The detailed virion structure of PnLV is unknown but the overall structure and particle stabilization of PnLV is expected to be similar to that of other viruses in the family (Aus dem Siepen et al., 2005). The primary amino acid sequence of PnLV is 17–30% identical with sobemovirus CPs, equivalent to the identities observed between sobemovirus CPs. Virus-like particles (VLPs) of potato leafroll virus (PLRV; genus Polerovirus) assembled from capsid protein subunits yield an icosahedral structure 30 nm in diameter at a resolution of 3.4 Å, that could be used for the modelling of the pea enation mosaic virus 1 (PEMV1, genus Enamovirus) VLP structure (Byrne et al., 2019). Structural superposition of the PEMV1 model with the respective experimentally determined structure of PLRV yielded RMSD of 0.84 Å. Analysis of structure similarities indicates that PLRV and PEMV1 group with the picorna-like capsid lineage, along with sobemoviruses (Byrne et al., 2019). Solemovirus icosahedral virions are comprised of 180 monomers of viral CP on a T=3 lattice symmetry. CP monomers are chemically identical but can adopt three conformations designated as A, B and C subunits. The A subunits form 12 pentamers at 5-fold axes, whereas three of each B and C subunits assemble into 20 hexamers at 3-fold axes (Figure 1. Solemoviridae) (Byrne et al., 2019, Opalka et al., 2000). The assembly of the T=3 capsid is controlled by the N-termini (known as random domain) of C subunits, which are partly ordered and inserted between the interacting sides of the subunits. The N-terminal arms of A and B subunits are completely disordered. The C-terminal shell domain forms a canonical single jellyroll β-sandwich fold (Rossmann et al., 1983). Whereas sobemovirus virions are stabilized by divalent cations Ca2+ and Mg2+ (Hull 1977), no metal ions were seen in PLRV VLP, and no putative metal binding sites were identified using bioinformatics (Byrne et al., 2019). With a low frequency, the virions of poleroviruses and enamoviruses incorporate coat protein subunits having the C-terminal extension of a readthrough protein associated with aphid transmission and virus particle stability (Bahner et al., 1990). The readthrough domain of CP was not involved in the structure refinement study (Byrne et al., 2019). Upon infection, a co-translational genome release happens by opening of the capsid at the tips of pentamers, whereas translation does not need complete disassembly of virions (Shields et al., 1989, Zink and Grubmüller 2010).
|
| Figure 1. Solemoviridae. Three-dimensional cryo-electron reconstruction of the particle of rice yellow mottle virus at 2.8 Å resolution. The capsid comprises 180 copies of a single capsid protein arranged in T=3 quasi-equivalent symmetry. The icosahedral asymmetric unit contains three subunits: A (in green), B (in brown), and C (in blue). Image from the RCSB PDB (rcsb.org) of PDB ID 1F2N (Opalka et al., 2000). |
Nucleic acid
The genome comprises a polycistronic, positive-sense, RNA of 4–6 kb. The 3′-terminus is non-polyadenylated but has a stable stem-loop or tRNA-like structure (Aus dem Siepen et al., 2005, Sõmera et al., 2015, Osman et al., 2006). Five sobemoviruses (rice yellow mottle virus, lucerne transient streak virus, subterranean clover mottle virus, Solanum nodiflorum mottle virus and velvet tobacco mottle virus) and one polerovirus (cereal yellow dwarf virus RPV) are known to encapsidate a small circular viroid-like satellite RNA (220–390 nt) (Symons and Randles 1999, Song and Miller 2004). Poleroviral and enamoviral capsids may also contain umbravirus RNA or a linear satellite RNA (717–902 nt) replicated by umbraviruses in co-infected plants (Demler et al., 1996, Menzel et al., 2009). In addition, polerovirus infection is commonly associated with tombusvirid-like RNAs of about 3 kb; these RNAs replicate autonomously but are dependent on polerovirus for vector transmission and intra-host systemic movement (Campbell et al., 2020).
Proteins
The capsid is comprised of 180 monomers of a single jellyroll capsid protein (CP) of 26–33 kDa in polemoviruses and sobemoviruses (Aus dem Siepen et al., 2005, Opalka et al., 2000) and 20–25 kDa in enamoviruses and poleroviruses (Alexander et al., 2017) . Occasional readthrough of an amber stop codon of the coat protein gene in enamoviruses and poleroviruses leads to translation of a 25–50 kDa C-terminal extension. This readthrough domain (RTD) protein is associated with aphid transmission, interaction with aphid endosymbiont proteins and virus particle stability (Brault et al., 1995, Hogenhout et al., 2000), and is needed for systemic infection (Boissinot et al., 2014, Xu et al., 2018). Virus particle formation is important for vector transmission (Musser et al., 2002, Kaplan et al., 2007), phloem transportation of poleroviruses (Hipper et al., 2014), xylem transportation of several sobemoviruses (Opalka et al., 1998) and their stability in the environment (Rossmann et al., 1983).
VPg is covalently attached to the 5′-terminus of genomic and subgenomic RNAs (Hacker and Sivakumaran 1997, Olspert et al., 2011, Olspert et al., 2011). Enamovirus VPg is also attached to the 5′-terminus of a umbravirus RNA and suggested to initiate umbravirus RNA encapsidation into the enamovirus capsid (Skaf et al., 2000).
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
Genome organization varies slightly between members of the four genera in the number of ORFs (4–10), and in the presence (enamoviruses, poleroviruses) or absence (polemovirus and sobemoviruses) of internal non-coding regions in addition to the short ones at the 5′- and 3′-ends (Figure 2. Solemoviridae). Viruses of one genus share the same basic genome structure. It is presumed that the gene function and expression strategy identified for viruses belonging to one species will apply to all others within the genus. All solemoviruses have a non-conserved 5′-proximal ORF (known as ORF0 in poleroviruses, polemoviruses and enamoviruses, and ORF1 in sobemoviruses) that encodes an RNA silencing suppressor (Sõmera et al., 2015, Csorba et al., 2015). An additional small non–conserved non-AUG initiated ORFx has been predicted in several sobemovirus genomes (Ling et al., 2013). The next ORF (ORF1 in poleroviruses, polemoviruses and enamoviruses, and ORF2a in sobemoviruses) encodes a polyprotein which consists of the motifs characteristic of a membrane-anchoring domain, a serine protease, a VPg, and the C-terminal domain. An RNA-directed RNA polymerase (RdRP) is translated via −1 programmed ribosomal frameshift (−1 PRF) from the next ORF (ORF2 in polerovirus, polemovirus and enamovirus genomes, and ORF2b in sobemovirus genomes) after the translation of the VPg domain as a part of longer polyprotein (Sõmera et al., 2015). These ORFs are translated from genomic RNA using a ribosomal leaky scanning strategy (Mayo et al., 1989, Sivakumaran and Hacker 1998). The −1 PRF signal of sobemoviruses consists of a slippery sequence 5′-UUUAAAC-3′ followed by a stem–loop structure, whereas the polemovirus frameshifting signal consists of the slippery sequence 5′-GGGAAAC-3′ followed by a putative small pseudoknot, this arrangement being similar to that observed for poleroviruses and enamoviruses (Aus dem Siepen et al., 2005, Tamm et al., 2009). The slippery heptanucleotide variations 5′-GGGAAAU-3′, 5′-UUUAAAC-3′or 5′-UUUAAAU-3′ can also be found in small number of polerovirus and enamovirus genomes (Prüfer et al., 1992, Miller and Giedroc 2010, Cao et al., 2019). Both versions of polyproteins are cleaved to different functional intermediates or definite subunits by the viral protease at E/R, E/S, E/T or E/N sites, there being up to four cleavage sites in the shorter polyprotein and up to three cleavage sites in the longer polyprotein (Li et al., 2000, Li et al., 2007, Mäkinen et al., 2000, Nair and Savithri 2010). The genome of PLRV contains an internal ribosomal entry site for a small unique ORF8 that encodes a replication-associated protein Rap1 expressed from the genomic RNA (Jaag et al., 2003).
The organization of the 3′-proximal half of solemovirus genomes is more diverse, indicating their different phylogenetic origin (Figure 2. Solemoviridae). These ORFs are translated from subgenomic RNA(s) using a ribosomal leaky scanning strategy (Tacke et al., 1990). For sobemoviruses and polemoviruses the capsid protein (CP) encoded by their ORF3 is the only protein expressed from sgRNA (Sõmera et al., 2015). Poleroviruses and enamoviruses encode several additional ORFs expressed from sgRNA1, sgRNA2 or sgRNA3. The major CP is encoded by ORF3 of sgRNA1, but if an in-frame readthrough of the amber stop codon occurs, translation is enabled to continue from ORF5 to produce the fusion protein CP-RTD. Translational readthrough of the stop codon is facilitated by a long-distance interaction with a downstream stem-loop structure (Xu et al., 2018). The polerovirus RTD is needed both for aphid transmission and for phloem retention (Peter et al., 2009). The sgRNA1 of poleroviruses expresses also two different movement proteins P3a and MP from ORF3a and ORF4, respectively. ORF3a is upstream of ORF3 and its translation initiates at a non-AUG codon. The ORF3a product is needed for establishment of systemic infection (Smirnova et al., 2015, Zhang et al., 2018). ORF4 is embedded within ORF3 in a different reading frame, and encodes a movement protein required for cell-to-cell movement (Lee et al., 2002). These two movement proteins interact with each other and with host organelles to transport the virus to the cell periphery (DeBlasio et al., 2018). Cell-to-cell movement ability of PLRV MP is dependent on targeting to a choline transporter-like 1 protein in plasmodesmata (Kraner et al., 2017). For several poleroviruses, the presence of one or two additional sgRNA has been reported. PLRV proteins P6 and P7 encoded by ORF6 and ORF7 are translated from sgRNA2 whereas PLRV sgRNA3 also encodes ORF7 and is able to express P7. The products of these ORFs participate in the regulation of virus replication and transcription, respectively (Hwang et al., 2013).
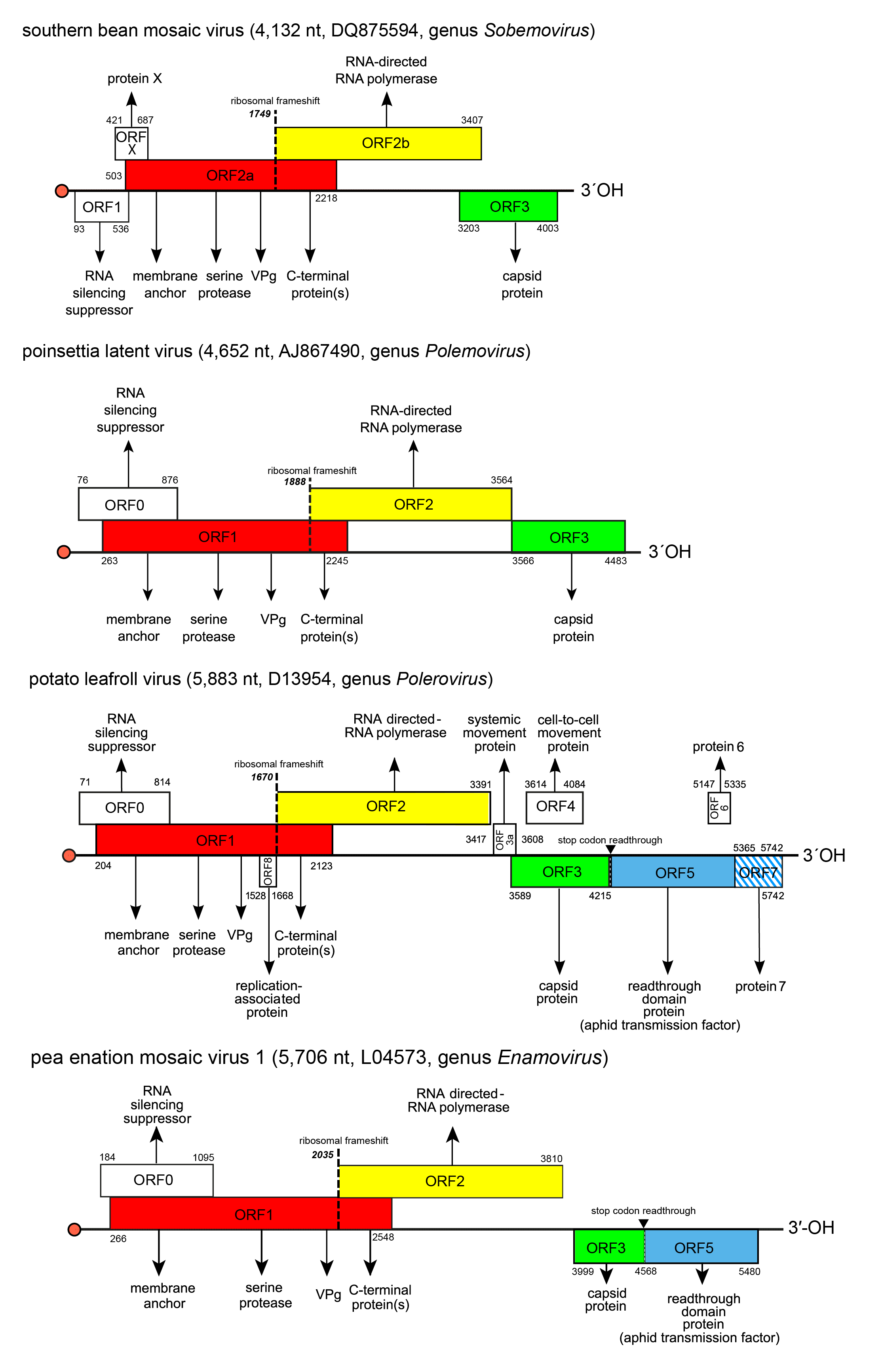 |
| Figure 2. Solemoviridae. Genome organization of the members of the genera Enamovirus, Polemovirus, Polerovirus and Sobemovirus. Red bullets indicate the virus protein genome-linked (VPg) covalently attached at the 5′-end of the RNAs. ORF reading frames are indicated by their position above and below the genome in the order -1, 0 and +1 and encode the polyprotein composed of the transmembrane domain, VPg, serine protease, and C-terminal protein(s) (red), RdRP (yellow). 3′-proximal ORFs encode the coat protein (CP, green) and readthrough domain protein related to aphid transmission properties (blue). Other ORFs have no sequence homology at the family level. ORF1 of sobemoviruses and ORF0 of enamoviruses, polemoviruses, and poleroviruses encode viral RNA silencing suppressors. Proteins encoded by ORFX of sobemoviruses and ORF3a of poleroviruses are needed for systemic infection. ORF4 of poleroviruses encodes movement protein, ORFs 6 and 7 encode proteins regulating the virus transcription and replication. ORF8 encodes a replication-associated protein that is unique for PLRV. The hatched area indicates an overlap between two ORFs in the same reading frame. The switch to ORF2 (or ORF2b in sobemoviruses) is determined by a −1 ribosomal frameshift (dashed line). ORFs 3a to ORF7 are expressed from subgenomic RNAs. |
Replication occurs after RdRP synthesis. A conserved 5′-end sequence 5′-ACAA(AA)-3′ and the 3′-end stem-loop or tRNA-like structure are considered to be essential for template recognition (Aus dem Siepen et al., 2005, Sõmera et al., 2015). The cellular compartments where the replication takes place are unknown, but membrane-anchoring is necessary (Osman et al., 2006, Govind et al., 2012). The VPg of the polerovirus cereal yellow dwarf virus RPV has shown to act as a primer for full-length negative-sense strand RNA synthesis on the positive-sense strand RNA template to form a dsRNA product (Osman et al., 2006). In vitro primer-independent replication has been demonstrated for the sobemovirus Sesbania mosaic virus – it has been proposed that the ACAA motif at the 3′-end of the negative-sense strand might act as promoter or enhancer for replicase binding and initiation of progeny RNA synthesis (Govind et al., 2012, Govind and Savithri 2010). Sobemoviruses, polemoviruses and enamoviruses transcribe one sgRNA, whereas poleroviruses generate up to three sgRNAs.
Polerovirus and sobemovirus VPgs have been demonstrated to interact with eukaryotic translation initiation factors eIF4 or eIF(iso)4G1 (Hébrard et al., 2010, Reinbold et al., 2013). In addition, sobemovirus VPg interacts with viral suppressor of RNA silencing and it regulates the activity of viral protease and viral RdRP (Govind et al., 2012, Roy Chowdhury and Savithri 2011, Nair et al., 2008). Multiple functions are supported by its intrinsically disordered nature with the propensity to form more rigid structures upon stabilization (Satheshkumar et al., 2005, Hébrard et al., 2009).
Biology
Solemoviruses are important pathogens of many major crops, including legumes, small grains, brassicas, sugarcane, potato and cucurbits. Viruses with high economic importance are rice yellow mottle virus, and papaya lethal yellowing virus among sobemoviruses; PLRV, sugarcane yellow leaf virus and several beet-infecting viruses among poleroviruses; and PEMV1 among enamoviruses.
Solemoviruses infect flowering plants from different botanical families, including both dicotyledons and monocotyledons. However, the natural host range of each solemovirus is narrow except that sowbane mosaic virus (Sobemovirus) and beet western yellows virus (Polerovirus) can both infect plants from several different orders.
Sobemoviruses are readily transmitted mechanically and many are also transmitted by insects such as beetles, aphids, hoppers and mirid bugs, this probably representing non–specific mechanical transmission by their mouthparts, although abiotic transmission has also been demonstrated (Sõmera et al., 2015). The polemovirus PnLV is transmitted via grafting and vegetative propagation, although virus infections in virus-free plants grown under quarantine conditions suggest that soil and water transmission may also occur (Aus dem Siepen et al., 2005). Abiotic transmission through soil or water is also highly likely for several sobemoviruses. Viruses that infect plants belonging to the families Fabaceae or Chenopodiaceae can be seed-transmissible (Sõmera et al., 2015). Sobemovirus infections range from symptomless to severe diseases (Aus dem Siepen et al., 2005, Sõmera et al., 2015, Trovão et al., 2015, Nascimento et al., 2010). Sobemovirus infection symptoms are described as mosaic and mottling in infected leaves, necrotic lesions, vein clearing, stunting and sometimes sterility. Sobemoviruses colonize leaf parenchymal tissues as well as the vascular tissues of leaves, stems and roots. Their particles have different preferences for vascular movement pathways: several of them move using the xylem whereas others spread through the phloem (Opalka et al., 1998, Otsus et al., 2012).
Polerovirus infections are often characterized by phloem necrosis, yellowing or reddening, streaking, rolling and thickening of the leaves and stunting of the plants. Enamovirus symptoms include stunting of the plant, chlorosis, puckering and vein enations on abaxial leaf surface. Poleroviruses and enamoviruses are phloem-specific. However, the phloem-restriction of enamoviruses is usually rescued by symbiotic umbraviruses, and inoculums made of the plant material with mixed infections are readily transmitted mechanically (Doumayrou et al., 2016). Also, mixed infections of poleroviruses and umbraviruses or potyviruses have been demonstrated to egress the phloem tissue and be readily transmitted by mechanical means (Barker 1989, Mayo et al., 2000). Such infections may be synergistic and show enhanced symptoms (Wintermantel 2005, Zhou et al., 2017). Mixed infections of poleroviruses or enamoviruses with umbraviruses may have a third component, a satellite RNA that is dependent on the polerovirus or enamovirus for encapsidation and vector transmission, and on the umbravirus for replication and systemic movement. The presence of a satellite RNA may aggravate umbravirus symptoms (Demler et al., 1996, Menzel et al., 2009).
In sobemovirus infected cells, virions are found in both cytoplasm and nucleus. Progression of infection leads to the appearance of large crystalline aggregates and inclusions in the cytoplasm and the vacuoles. Changes in chloroplast structure have been reported for several infections (Sõmera et al., 2015).
In polerovirus-infected phloem cells, virions are associated with virus-induced vesicles in the cytoplasm, these often observed fusing with the nucleus, mitochondria, vacuoles, and ER membranes associated with plasmodesmata (Esau and Hoefert 1972, Shepardson et al., 1980).
Whereas sobemoviruses are mainly transmitted via mechanical wounds (Sõmera et al., 2015), transmission of poleroviruses and enamoviruses is by particular aphid species in a persistent, circulative and non-propagative manner, i.e. these viruses enter the hemocoel of the aphid gut by a receptor-mediated transport process, circulate in the hemolymph and enter the accessory salivary gland by a second receptor-mediated transport event (Brault et al., 2007, Boissinot et al., 2017). Movement across tissue barriers within the gut and accessory salivary glands occurs via clathrin-mediated endocytosis (Gray and Gildow 2003). Observations indicate that infected plants are more attractive to aphid vectors than are healthy plants, and the aphid vectors often develop more rapidly and produce more offspring on infected hosts. Such differences have been linked to the difference in plant volatile organic compounds composition in infected and non-infected plants (Ziegler-Graff 2020). In PLRV-infected plants, increased aphid attraction has been correlated with a reduction of jasmonic acid and ethylene signalling compared to uninfected control plants (Patton et al., 2020).
Antigenicity
Virions and purified CPs are strongly immunogenic. Distant serological relationships exist between the CPs of some sobemoviruses or between the CPs or MPs of poleroviruses. Serotypes with different geographical origin have been identified for some viruses. No serological relationships have been reported between sobemoviruses and poleroviruses or enamoviruses. Heterologous serological crossreactions have sometimes been detected between the CPs of poleroviruses and luteoviruses CPs, reflecting their common phylogenetic origin (D’Arcy et al., 1989). In gel diffusion assays, polerovirus aphid-transmissible isolates sometimes display antigenic determinants that are absent from aphid-non-transmissible isolates. Serological relationships may be detected when comparing disrupted virus particles that are not detectable when intact virions are tested.
Derivation of names
Enamovirus: from the species Pea enation mosaic virus.
Polemovirus: from the genus names Polerovirus and Sobemovirus to indicate the chimeric nature of the polemovirus genome.
Polerovirus: from the species Potato leafroll virus.
Sobemovirus: from the species Southern bean mosaic virus.
Solemoviridae: from the “founding” genus names Sobemovirus and Polemovirus to indicate the close phylogenetic relationship of solemoviruses resulting from recombination during evolution.
Genus demarcation criteria
The genera in the family Solemoviridae are differentiated by phylogenies of individual ORFs or proteins. Structural comparisons of encoded proteins or virions may help to resolve phylogenetic relationships.
Relationships within the family
The RNA silencing suppressors encoded by the 5′-proximal ORF of solemoviruses are generally not conserved (except the zinc-finger like motifs of sobemovirus P1s and the F-box-like motifs of polemovirus, polerovirus and enamovirus P0s) and share no significant homology with any other known proteins (Aus dem Siepen et al., 2005, Sõmera et al., 2015, Fusaro et al., 2012, Rashid et al., 2019).
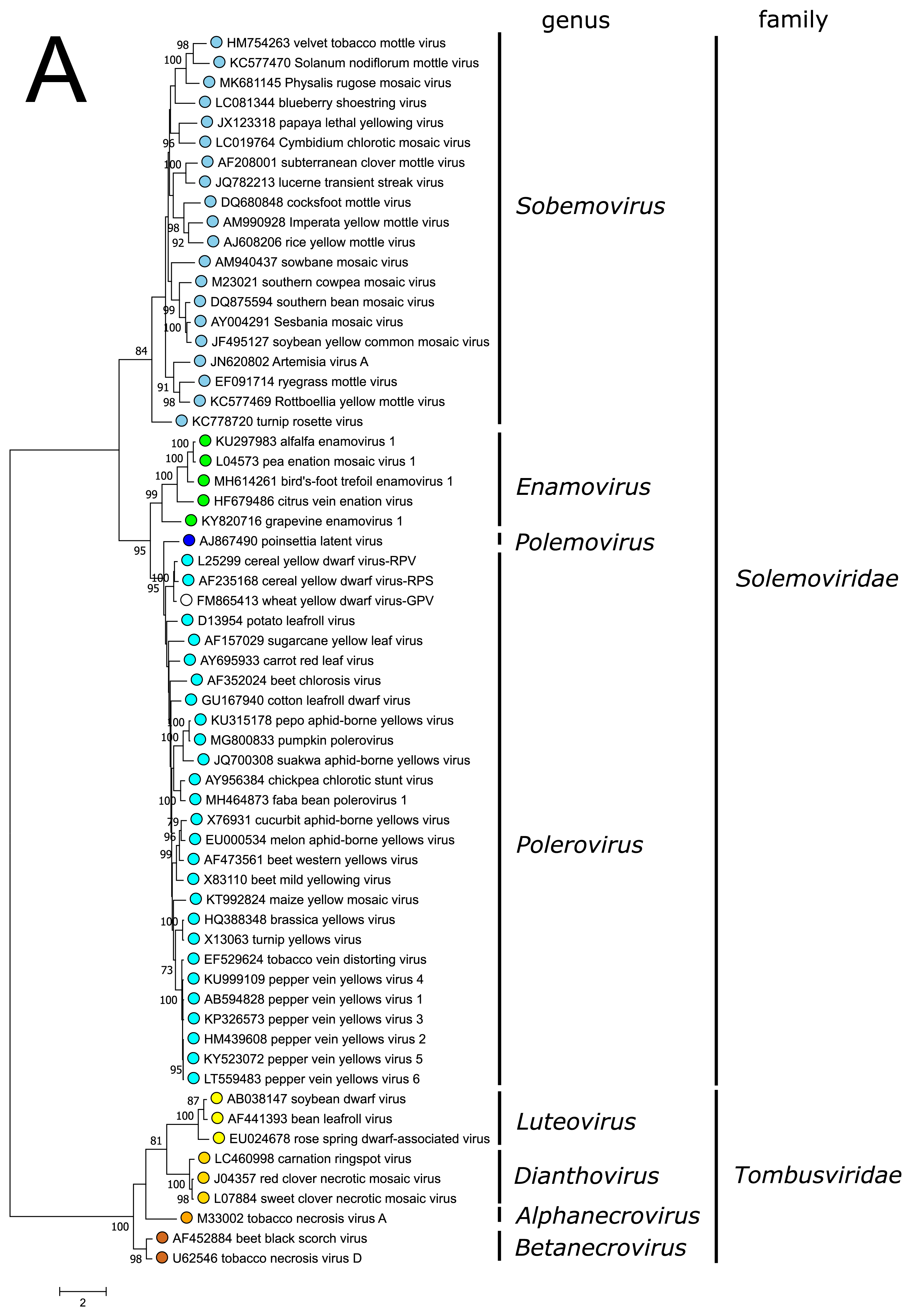
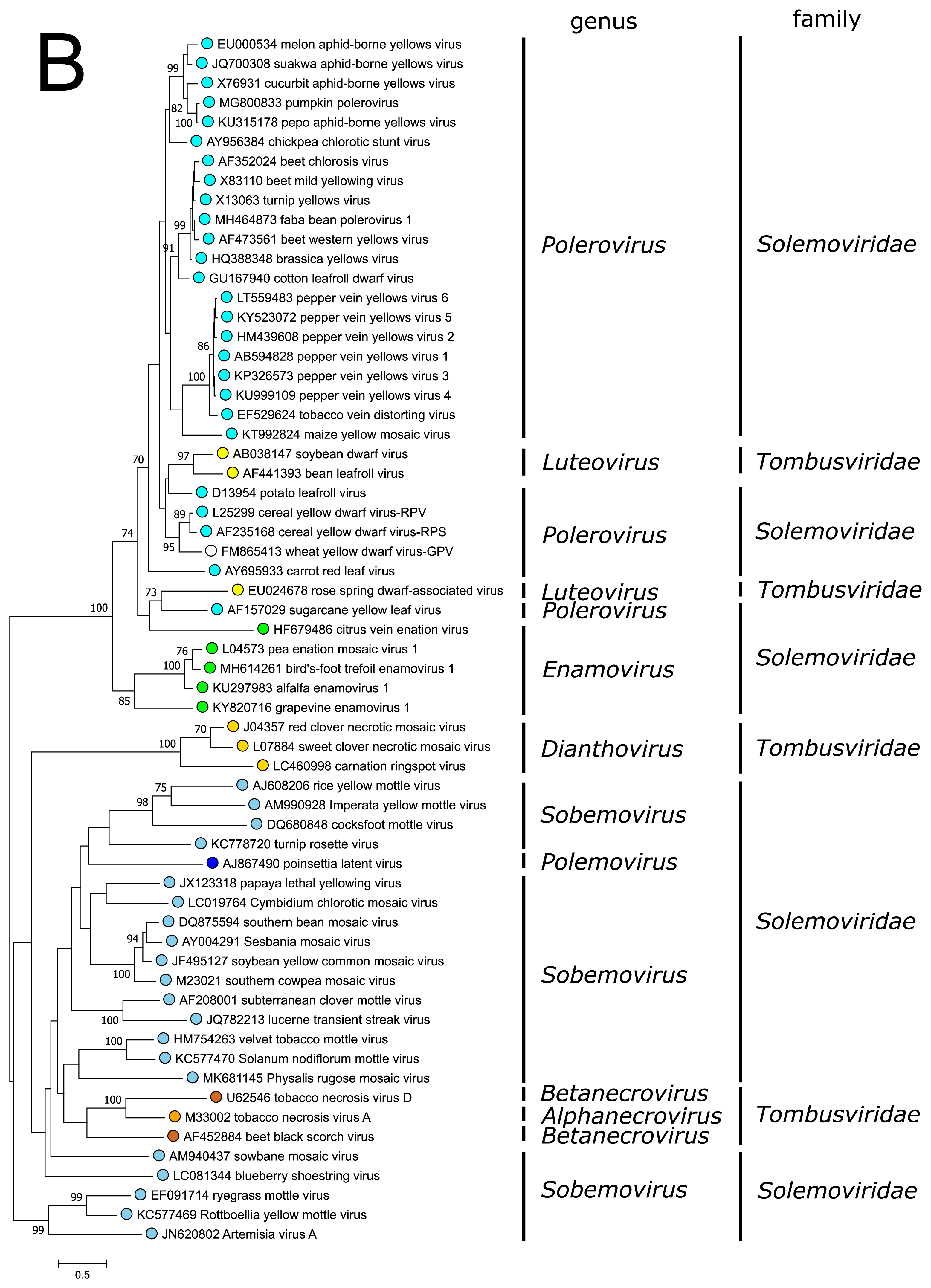
The polyprotein Pro–VPg–RdRP of the polemovirus PnLV shares the highest sequence identities with poleroviruses (Aus dem Siepen et al., 2005). Respectively, PnLV RdRP groups with that of poleroviruses, which are phylogenetically closer to the enamovirus RdRPs than to the sobemovirus RdRPs (Figure 3A).
PnLV is considered as a natural polerovirus-sobemovirus recombinant (Aus dem Siepen et al., 2005). Indeed, close sequence relationships can be found between the PnLV CP and sobemovirus CPs. The group of sobemovirus CPs of Artemisia virus A (ArtVA), ryegrass mottle virus (RGMoV) and Rottboellia yellow mottle virus (RoMoV), are more distantly related to other sobemovirus CPs which is in concordance with studies on RGMoV virion structure showing that RGMoV has a relatively smaller particle diameter compared to other sobemovirus virions (Plevka et al., 2007). 3D homology modelling of the CP of RoMoV indicates the overall fold characteristic of RGMoV CP (Sõmera and Truve 2015). Similarly, the CPs of poleroviruses and enamoviruses are closely related to each other (Figure 3B), and these viruses also share the readthrough domain protein that is expressed as the C-terminal extension of CP if the amber stop codon of CP gene is suppressed.
|
|
| Figure 3. Solemoviridae. Maximum likelihood phylogenetic trees constructed using the amino acid sequences of (A) RNA-directed RNA polymerase domains (starting at a −1 ribosomal frameshifting site) and (B) capsid proteins. Phylogenetic analysis based on a MUSCLE multiple sequence alignment was performed with either LG (RdRP) or WAG (CP) amino acid substitution model using Seaview 5.0.4 (http://doua.prabi.fr/software/seaview). Both sets of sequences were translated from the complete genomes of the same isolate. The proportion of branches found in 500 bootstrap replicates is indicated if this was > 70%. Coloured dots group members of a genus, with an unclassified virus marked with an open circle. The examples of RdRPs and CPs of alphanecroviruses, betanecroviruses, dianthoviruses and luteoviruses representing the members of family Tombusviridae in the order Tolivirales were used to demonstrate their distant grouping from solemoviral RdRPs but closer relationships with their CPs, respectively. The phylogenetic trees and the corresponding sequence alignments are available for download at the Resources page. |
Relationships with other taxa
The modules in the genome of solemoviruses encoding Pro–VPg–RdRP or CP share sequence homology with the gene modules belonging to viruses of different taxa.
The RdRPs of viruses from family Solemoviridae share the closest phylogenetic relationships with the RdRPs of viruses belonging to the families Alvernaviridae and Barnaviridae in the order Sobelivirales (Wolf et al., 2018). All these viruses are assumed to encode their picornavirus-like RdRP as part of a long polyprotein produced via −1 programmed frameshift mechanism downstream of serine protease and VPg domains. The exception is Heterocapsa circularisquama RNA virus 01 belonging to the family Alvernaviridae for which the polyprotein is a product of translation from a single frame. Although less conserved, the serine protease sequences of these viruses are phylogenetically related to each other. Their VPg sequences are not well conserved but their position between viral protease and RdRP domains is unique among the phylogenetically related sobemoviruses, polemoviruses, poleroviruses, enamoviruses and barnaviruses (Sõmera et al., 2015).
The CPs of poleroviruses and enamoviruses have a common evolutionary origin with those of the members of genus Luteovirus in the family Tombusviridae (order Tolivirales) (Figure 3B), with which they also share the readthrough domain (RTD) that is expressed as the C-terminal extension when the amber stop codon of CP gene is suppressed. The CPs of polemoviruses and sobemoviruses are most closely related to the CPs of necroviruses belonging to the family Tombusviridae in the order Tolivirales, which cluster within sobemovirus CP lineage (Figure 3B) and show a 3D-virion structure close to sobemovirus 3D-virion structures (Plevka et al., 2007). The capsids of viruses belonging to the families Solemoviridae and Tombusviridae have significant structural similarities to those of members of the families Secoviridae, Tymoviridae and Astroviridae, belonging to the picorna-like capsid lineage recognized by the single jelly-roll capsid protein fold (Byrne et al., 2019).
Previous taxonomic classification of the genera Polerovirus, Enamovirus and Luteovirus placed them into the family Luteoviridae according to their CP/CP-RTD phylogeny. It has been suggested that the viruses belonging to the genera Polerovirus, Enamovirus and Luteovirus have been produced via recombinational events between an ancestral virus genome containing the capsid protein genes characteristic of these viruses, and virus genomes encoding a polymerase phylogenetically related either to that of sobemoviruses or dianthoviruses (Miller et al., 1997).
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Beihai sobemo-like virus 1 | KX882805 | BSLV1 |
| Ginkgo biloba sobemo-like virus | MN831440 | GbSLV |
| Hubei sclerotinia RNA virus 1 | MK889164 | HuSRV1 |
Virus names and virus abbreviations are not official ICTV designations.