Family: Alphaflexiviridae
Jan F. Kreuze, Anna Maria Vaira, Wulf Menzel, Thierry Candresse, Sergei K. Zavriev, John Hammond and Ki Hyun Ryu
The citation for this ICTV Report chapter is the summary published as Kreuze et al., (2020):
ICTV Virus Taxonomy Profile: Alphaflexivridae, Journal of General Virology, 101, 699–700
Corresponding author: Jan F. Kreuze ([email protected])
Edited by: F. Murilo Zerbini and Stuart G. Siddell
Posted: April 2020
PDF: ICTV_Alphaflexiviridae.pdf
Summary
The family Alphaflexiviridae includes viruses with flexuous filamentous virions that are 470–800 nm long and 12–13 nm in diameter (Table 1.Alphaflexiviridae). Alphaflexiviruses have a single-stranded, positive-sense RNA genome of 5.4–9 kb. They infect plants and plant-infecting fungi. They share a distinct lineage of alphavirus-like replication proteins that is unusual in lacking any recognized protease domain. Cell-to-cell and long-distance movement is facilitated by triple gene block proteins in viruses that infect plants, except for members of the genus Platypuvirus.
Table 1.Alphaflexiviridae. Characteristics of members of the family Alphaflexiviridae
| Characteristic | Description |
| Typical member | shallot virus X, Kanyuka (M97264), species Allexivirus ecsascalonicum |
| Virion | Flexuous filaments, usually 12–13 nm in diameter and from 470 to about 800 nm in length |
| Genome | Single molecule of linear ss, positive sense RNA of about 5.5–9.0 kb |
| Replication | Cytoplasmic, virus-coded RdRP |
| Translation | Genome length and 3′-terminal subgenomic mRNAs; capped and polyadenylated |
| Host range | Plants and fungi |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Kitrinoviricota, class Alsuviricetes, order Tymovirales, six genera with 72 species |
Virion
Morphology
Virions are flexuous filaments, usually 12–13 nm in diameter (range 10–15 nm) and from 470 to about 800 nm in length. Alphaflexiviruses have helical capsid symmetry with a pitch of about 3.4 nm (range 3.3–3.7 nm) and in the members of some genera there is clearly visible cross-banding (Figure 1. .Alphaflexiviridae).
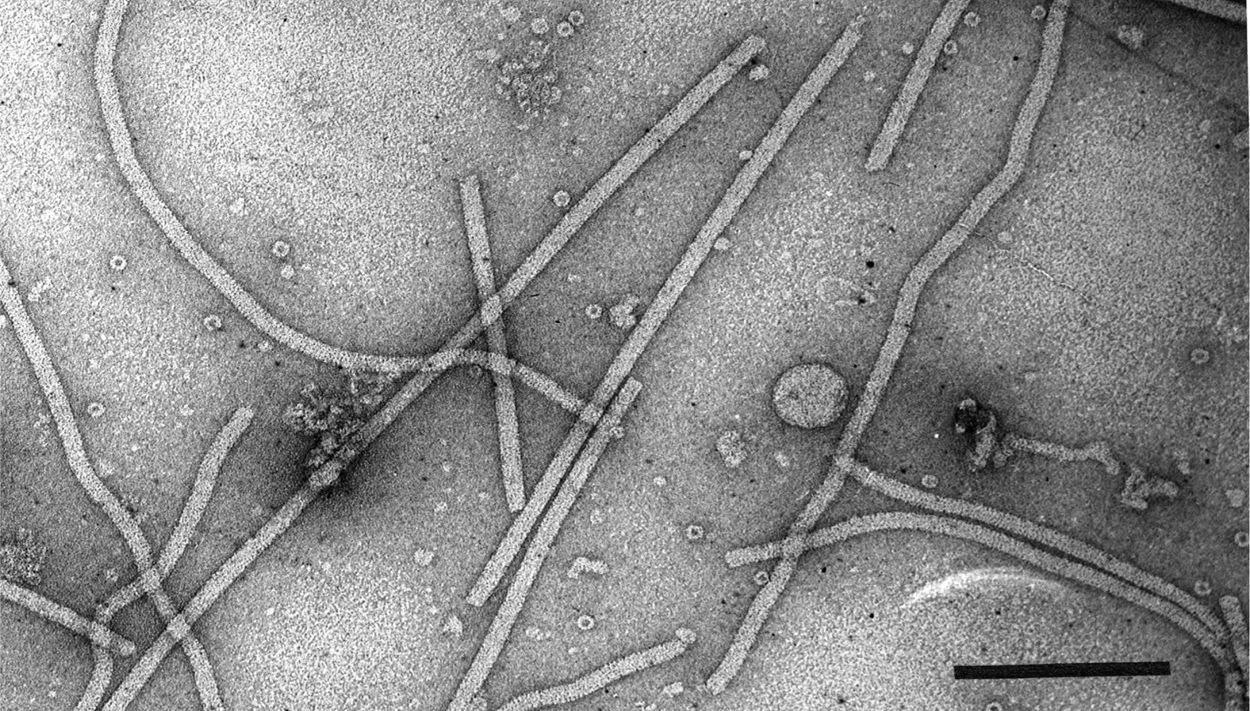 |
| Figure 1. .Alphaflexiviridae. Electron micrograph of negatively-stained virions of an isolate of shallot virus X. Bar = 200 nm. |
Physicochemical and physical properties
Virions sediment as a single band (or occasionally two very close bands) with an S20,w of 92–176S.
Nucleic acid
Virions contain a single molecule of linear single-stranded RNA of 5.9–9.0 kb which is 5–6% by weight of the virion. Where studied, the RNA is capped at the 5′-terminus with m7G and has a polyadenylated tract at the 3′-terminus (Figure 2. .Alphaflexiviridae). Open reading frames downstream from the large ORF encoding the replication-associated (Rep) protein are expressed from smaller 3′-co-terminal subgenomic RNAs (sgRNAs). These sgRNAs are encapsidated in some, but not all, members of the genus Potexvirus.
Proteins
The viral capsid is composed of a single polypeptide ranging in size from 18 to 43 kDa except for members of the genus Lolavirus which have two carboxy-coterminal capsid protein (CP) variants, and members of the genus Sclerodarnavirus in which no CP has been identified (Vaira et al., 2011). In allexiviruses, a 42 kDa polypeptide was also detected as a minor component of virions. In lolaviruses a shorter (ca. 28 kDa) carboxy co-terminal polypeptide forms an equimolar fraction of the virion with the polypeptide (ca. 32 kDa) originating from the first AUG of the CP-encoding ORF.
Lipids
None reported.
Carbohydrates
Usually none, but the capsid protein of some strains of the species Potato virus X (genus Potexvirus) and both forms of the lolavirus capsid protein are reported to be glycosylated.
Genome organization and replication
Most alphaflexiviruses have five to seven genes, but members of the genus Sclerodarnavirus have a single gene encoding the Rep protein (Figure 2. .Alphaflexiviridae). The ORF1-encoded product (encoded by ORF2 in members of the genus Platypuvirus), which follows a short 5′-UTR sequence, has homologies with replication-associated (Rep) proteins of the “alphavirus-like” supergroup of RNA viruses. This protein (150–195 kDa) contains conserved methyltransferase, helicase and RNA-directed RNA polymerase (RdRP) motifs (Batten et al., 2003); some also include an AlkB domain (alkylated DNA repair protein) (van den Born et al., 2008). In all plant-infecting alphaflexviruses, except members of the genus Platypuvirus, ORFs 2–4 encode the “triple gene block” (TGB) proteins involved in cell-to-cell movement and ORF5 is the viral capsid protein. In platypuviruses the CP is encoded by ORF 4 and a putative movement protein (MP) is found at ORF7. In members of some genera (Allexivirus except alfalfa virus S, Lolavirus and Mandarivirus) a final ORF encodes a protein with a zinc-finger motif and the ability to bind nucleic acids. ORFs downstream of the polymerase are translated from 3′-terminal sgRNAs that can often be found in infected tissue. Replication is known, or presumed to be, cytoplasmic and the product of ORF1 is the only virus-encoded protein known to be involved.
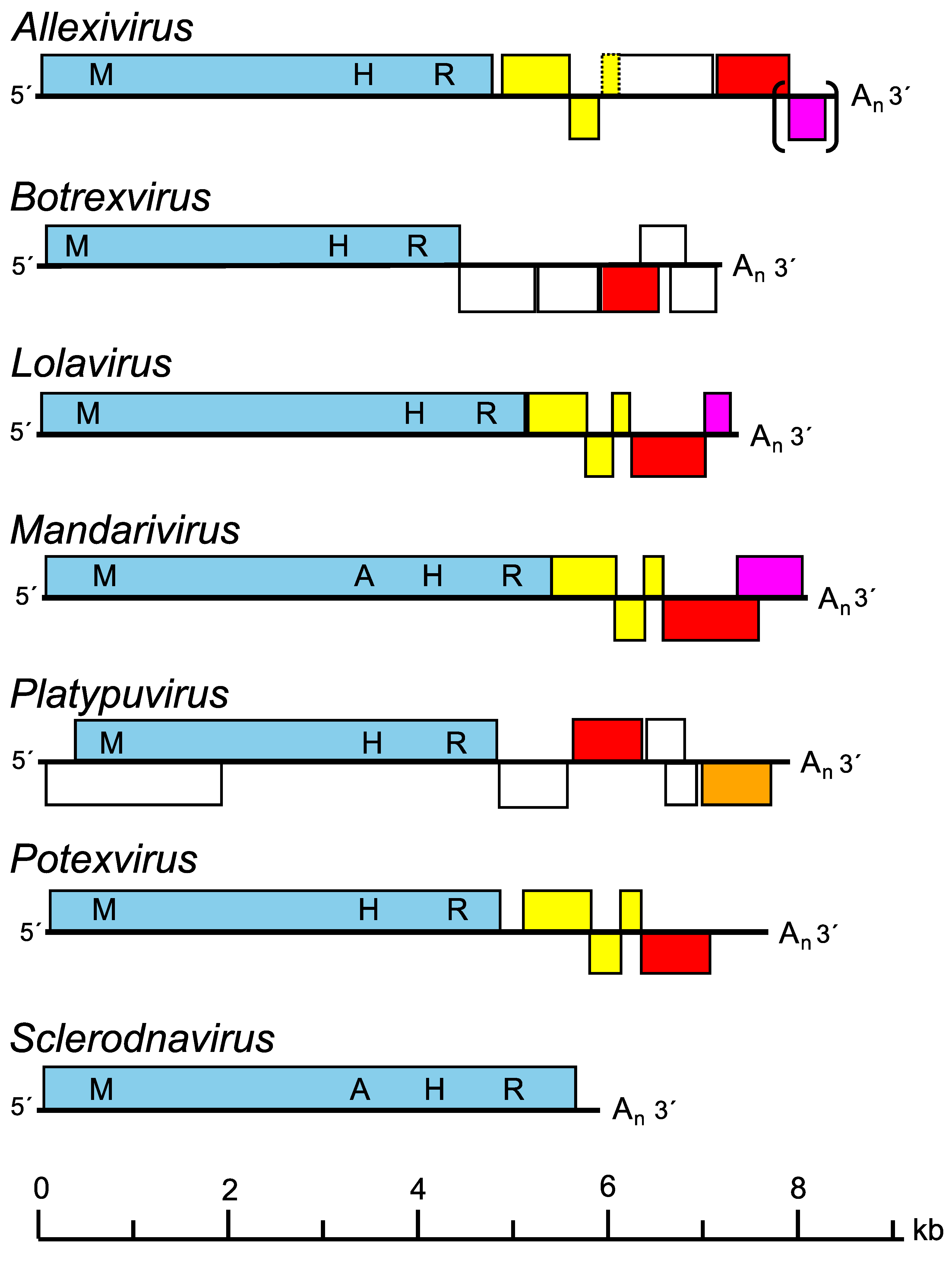 |
| Figure 2. .Alphaflexiviridae. Diagram showing genome organization of members of genera in the family Alphaflexiviridae. Blocks represent predicted open reading frames (ORFs). Rep proteins are shown in blue, triple gene block (TGB) proteins in yellow, 3A-like movement protein in orange, capsid proteins in red and RNA binding proteins in purple. Other ORFs are white. The methyltransferase (M), AlkB (A), helicase (H) and RNA-directed RNA polymerase (R) domains of the replicase are also shown. Brackets indicate ORFs may be missing from some members of a genus and the dashed outline of an ORF indicates that the start codon is missing in virus isolates of several species of the genus. |
Biology
Alphaflexiviruses have been reported from a wide range of mono- and dicotyledonous plant species but the host range of members of individual species is usually limited. Many viruses have relatively mild effects on their host. All plant-infecting viruses can be transmitted by mechanical inoculation, often readily. Many viruses have no known invertebrate or fungal vectors; however, some allexiviruses are mite-borne. Aggregates of virus particles accumulate in the cytoplasm but there are usually no specific cytopathic structures.
Antigenicity
Virions are usually highly immunogenic. Some viruses within genera are serologically related.
Derivation of names
Alpha: from Greek letter ɑ
Flexi: from Latin flexus, meaning “bent”
Genus demarcation criteria
Members of different genera are distinguished by various features of genome organization (Figure 2. .Alphaflexiviridae) and host (Table 2.Alphaflexiviridae). Viruses from different genera usually have less than 45% nt identity in the CP or polymerase genes.
Table 2.Alphaflexiviridae. Characteristics of members of the seven genera in the family Alphaflexiviridae
| Genus | Host | Virion length (nm) | ORFs | Rep (kDa) | MP | CP (kDa) |
| Allexivirus | plants | ca. 800 | 5–6 | 170–195 | TGBa | 25–32 |
| Botrexvirus | fungi | ca.720 | 5 | 158 | Absent | 43 |
| Lolavirus | plants | 640 | 6 | 196 | TGB | 28, 32 |
| Mandarivirus | plants | 650 | 6 | 187 | TGB | 34 |
| Platypuvirus | plants | No virions | 7 | 157 | 3A-like | 22 |
| Potexvirus | plants | 470–580 | 5 | 150–195 | TGB | 21–27 |
| Sclerodarnavirus | fungi | No virions | 1 | 193 | Absent | Absent |
a TGB, triple gene block
Relationships within the family
In a phylogenetic analysis of the Rep protein of isolates for which a CP sequence was also available, members of most genera fall into well-supported clades, except for members of the genus Mandarivirus which group with members of the genus Potexvirus (Figure 3. .Alphaflexiviridae). A similar tree is found based on CP sequences except that members of the genus Lolavirus group together with members of the genera Mandarivirus and Potexvirus (Figure 4. .Alphaflexiviridae) and may be the result of modular recombination during evolution of viruses belonging to these genera.
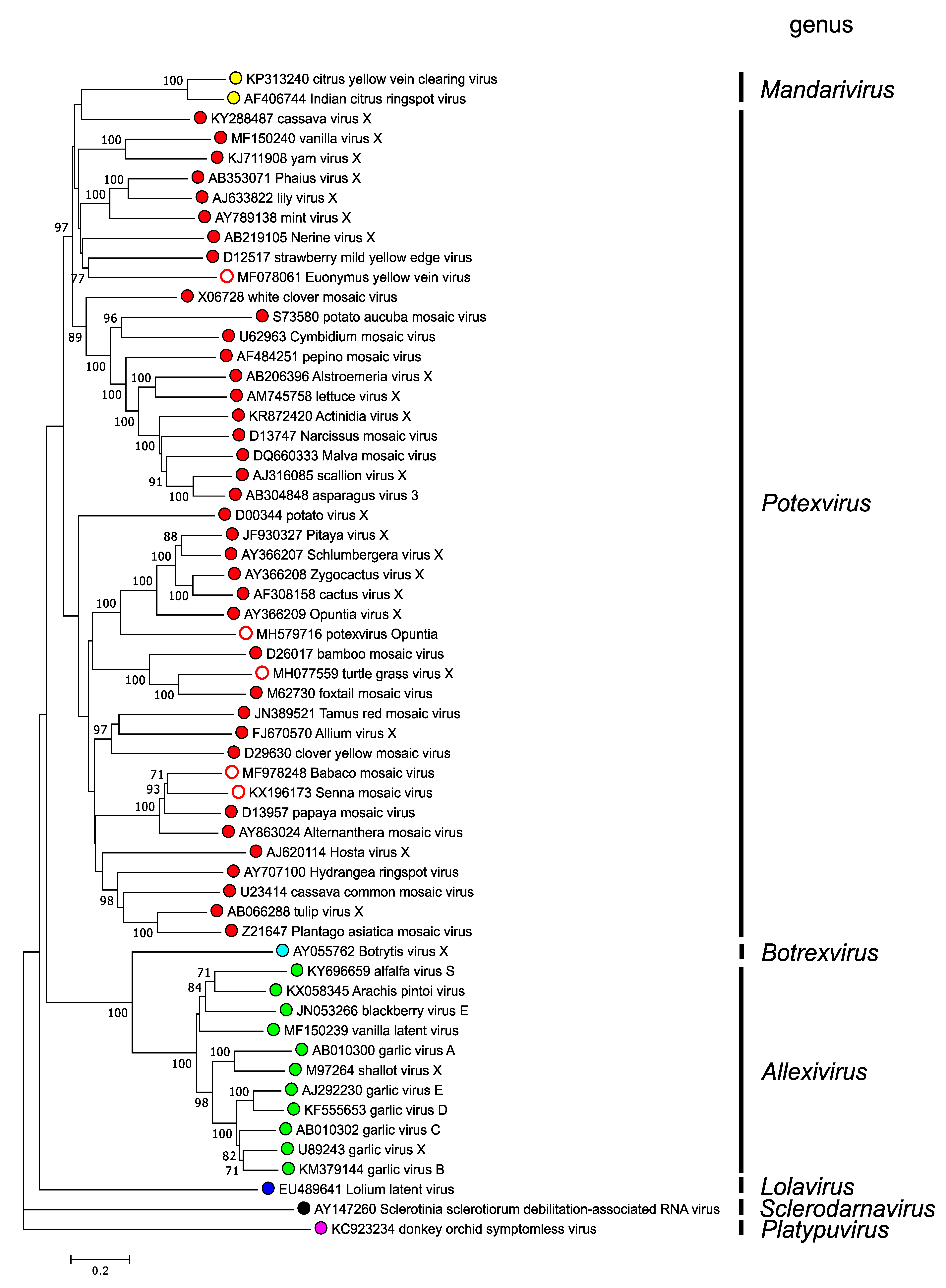 |
| Figure 3. .Alphaflexiviridae. Unrooted phylogenetic (distance) tree based on the amino acid sequences of the entire Rep protein of members of the family Alphaflexiviridae. Virus genera are indicated to the right of their respective branches. Numbers on branches indicate percentage of bootstrap support out of 1000 bootstrap replications (when >60%). The scale indicates JTT amino acid distances. Tree produced in (Tamura et al., 2007). This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
 |
| Figure 4. .Alphaflexiviridae. Unrooted phylogenetic (distance) tree based on the amino acid sequences of the capsid protein of members of the family Alphaflexiviridae. Virus genera are indicated to the right of their respective branches. Numbers on branches indicate percentage of bootstrap support out of 1000 bootstrap replications (when >70%). The scale indicates JTT amino acid distances. Tree produced in MEGA7 (Kumar et al., 2016). This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Relationships with other taxa
The Rep proteins are members of the “alphavirus-like” supergroup of RNA viruses and are most closely related to those encoded by members of other families in the order, namely Betaflexiviridae, Gammaflexiviridae, Deltaflexiviridae and Tymoviridae. The TGB proteins are related to those of members of some genera in the family Betaflexiviridae and, more distantly, to those of rod-shaped viruses in the family Virgaviridae (genera Hordeivirus, Pecluvirus and Pomovirus). In members of the genus Platypuvirus, ORF1 shows weak similarity to tymovirus movement proteins and the ORF7 movement protein shows similarity to those encoded by members of the families Tombusviridae and Virgaviridae.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Ambrosia asymptomatic virus 1 | KF421905 | |
| cassava alphaflexivirus | KC505252 | |
| Escobaria virus | KF421919 | |
| Euonymus yellow mottle associated virus | MK572000 | EuYMaV |
| grapevine associated alphaflexivirus-1 | HM852918 | |
| insect-associated alphaflexivirus1 | MN203143 | |
| insect-associated alphaflexivirus 2 | MN203144 | |
| insect-associated alphaflexivirus 3 | MN203145 |
Virus names and virus abbreviations are not official ICTV designations.

