Family: Baculoviridae
Genus: Deltabaculovirus
Distinguishing features
Only a single species (Deltabaculovirus cunigripalpi) has been classified in this genus. Replication of the isolate Culex nigripalpus nucleopolyhedrovirus (CuniNPV) is restricted to host midgut epithelium, primarily in larval stages but rarely in adults. Two virion phenotypes may be characteristic of a virus species. Virions of the occlusion-derived virus phenotype are embedded within an occlusion body composed of a crystalline matrix of a single viral protein with no homology to polyhedrin or granulin proteins of other baculovirus genera (Moser et al., 2001, Perera et al., 2006). Each occlusion body ranges in size from 0.5 to 5 µm and contains few (1–4) or many (>50) singly-enveloped virions depending on the strain of virus, lacks the polyhedron envelope of other baculoviruses, but also matures within nuclei of infected cells (Figure 1.Deltabaculovirus).
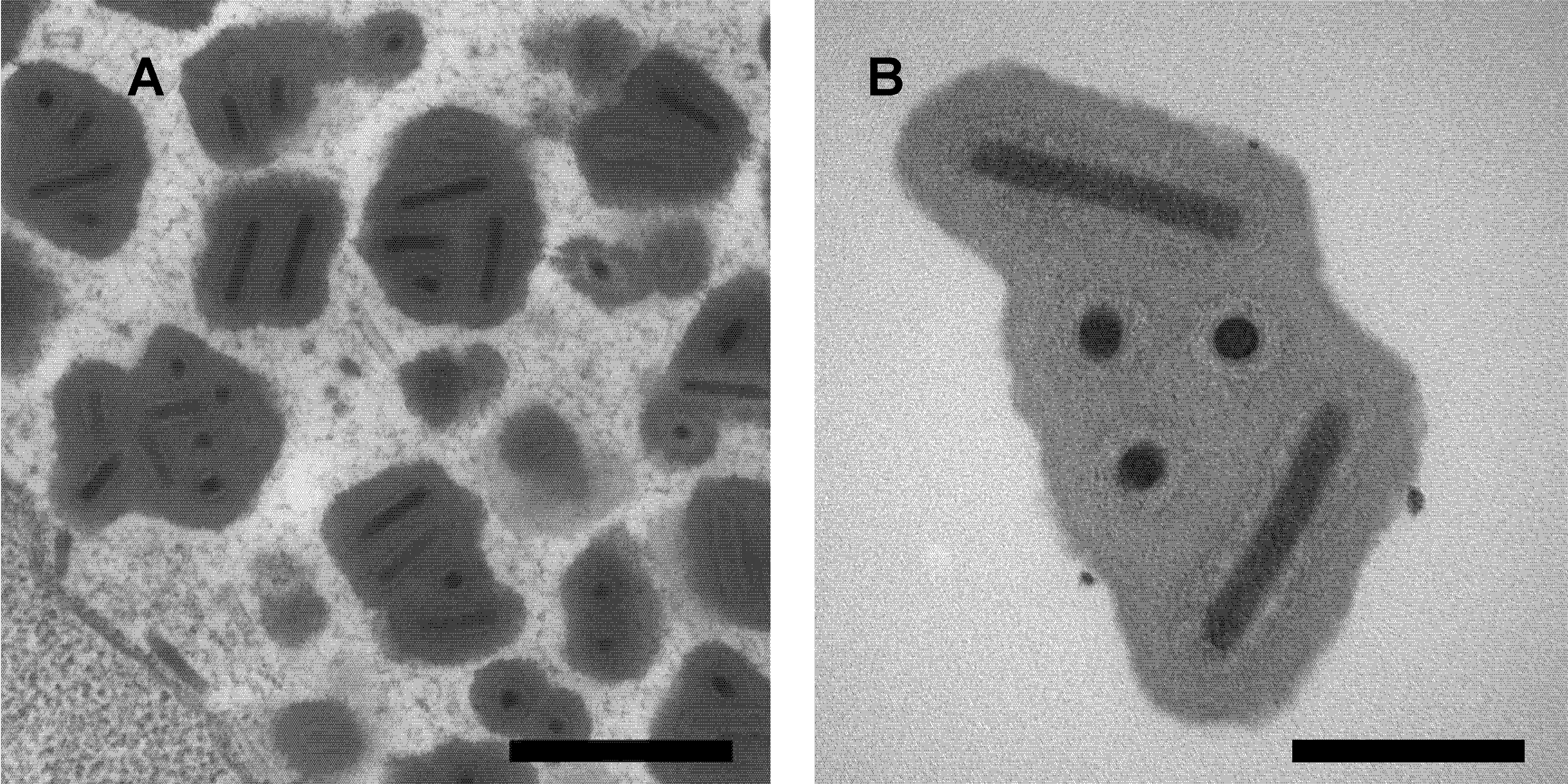 |
| Figure 1.Deltabaculovirus. Transmission electron micrographs of occlusion bodies of the deltabaculovirus Culex nigripalpus nucleopolyhedrovirus. Scale bars: (A) 0.5 µm; (B) 0.2 µm. |
Virion
See discussion under family properties.
Genome organization and replication
The CuniNPV genome is 108,252 bp and encodes at least 109 putative proteins, some of which have sequence homology with those from other baculoviruses (Afonso et al., 2001). Homologous proteins are involved in early and late gene expression, DNA replication, as well as structural and auxiliary functions. Gene orientation, order, and content in the genome of CuniNPV is different from the members of other baculovirus genera.
Biology
Transmission of CuniNPV to larval mosquitoes is strongly influenced by divalent cations: Mg2+ is a potent enhancer of transmission whereas Ca2+ is a strong inhibitor (Becnel et al., 2001). Hosts include at least three genera of mosquitoes, but other mosquito genera and families of Diptera are likely hosts.
Species demarcation criteria
Only a single species has been classified for this genus, although other isolates that likely represent new species have been reported (Becnel and White 2007). It is possible that criteria based on alignments of conserved gene sequences can be developed (de Araujo Coutinho et al., 2012).
Related, unclassified viruses
| Virus name | Accession number | Viurs abbreviation |
| Aedes sollicitans nucleopolyhedrovirus | JQ582836; JQ582837 | AesoNPV |
| Uranotaenia sapphrinia nucleopolyhedrovirus | JQ582838; JQ582839 | UrsaNPV |
Virus names and virus abbreviations are not official ICTV designations.

