Subfamily: Alpharhabdovirinae
Genus: Arurhavirus
Distinguishing features
Viruses assigned to the genus Arurhavirus form a distinct monophyletic group based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L protein sequences. Viruses assigned to the genus have been discovered in forest-dwelling mosquitoes and sandflies. There is evidence of infection in rodents and, possibly, birds. Arurhaviruses are most closely related to rhabdoviruses in the genus Curiovirus.
Virion
Morphology
Xiburema virus (XIBV; species Arurhavirus xiburema) virions have typical rhabdovirus morphology by electron microscopy (Karabatsos 1985).
Nucleic acid
Arurhavirus genomes consist of a single molecule of negative-sense, single-stranded RNA and range from 11.9–12.3 kb (Wanzeller et al., 2014, Walker et al., 2015).
Proteins
Arurhavirus N, P, M, G and L proteins share sequence homology and/or structural characteristics with the cognate proteins of other rhabdoviruses. Other proteins encoded in arurhavirus genomes have not yet been identified in infected cells. The putative U1 proteins of XIBV, Aruac virus (ARUV; species Arurhavirus aruac), and Inhangapi virus (INHV; species Arurhavirus inhangapi), and the Gx protein of Santa Barbara virus (SBAV; species Arurhavirus santabarbara), are class 1a viroporin-like proteins (11.1–14.1 kDa) featuring an N-terminal domain containing large hydrophobic residues, a central transmembrane domain and a highly basic C-terminal domain. The putative SBAV U1 protein (9.0 kDa), ARUV U1x protein (5.9 kDa) and XIBV U2 protein (7.8 kDa) are unique proteins of unknown function.
Genome organisation and replication
Arurhavirus genomes include five genes (N, P, M, G and L) encoding the structural proteins and all also encode a class 1a viroporin-like protein (Wanzeller et al., 2014, Walker et al., 2015) (Figure 1.Arurhavirus). In ARUV, INHV and XIBV, the viroporin-like protein is encoded in an additional transcriptional unit (gene) (U1) between the G and L genes; in SBAV, the viroporin-like protein is encoded in an alternative ORF (Gx) overlapping the end of the G ORF. In the ARUV U1 gene, encoding the viroporin-like protein, contains a second ORF (U1x) of 156 nt. The U1 termination codon and the U1x initiation codon overlap and are preceded by a TURBS (termination upstream ribosome-binding site)-like sequence (5′-AGGGA-3′) suggesting that expression of ORF U1x occurs by a stop-start mechanism as observed for similar arrangements in several other rhabdoviruses, including ephemeroviruses, hapaviruses, curioviruses and sripuviruses. The XIBV U1 gene, encoding the viroporin-like protein, is followed by a gene (U2) that contains a small ORF of 65 nt. There is no significant sequence homology between the ARUV U1x and XIBV U2 proteins. The SBAV genome, uniquely amongst the arurhaviruses, contains an additional long ORF in an independent gene (U1) between the N and P genes, encoding a 9.0 kDa protein of unknown function.
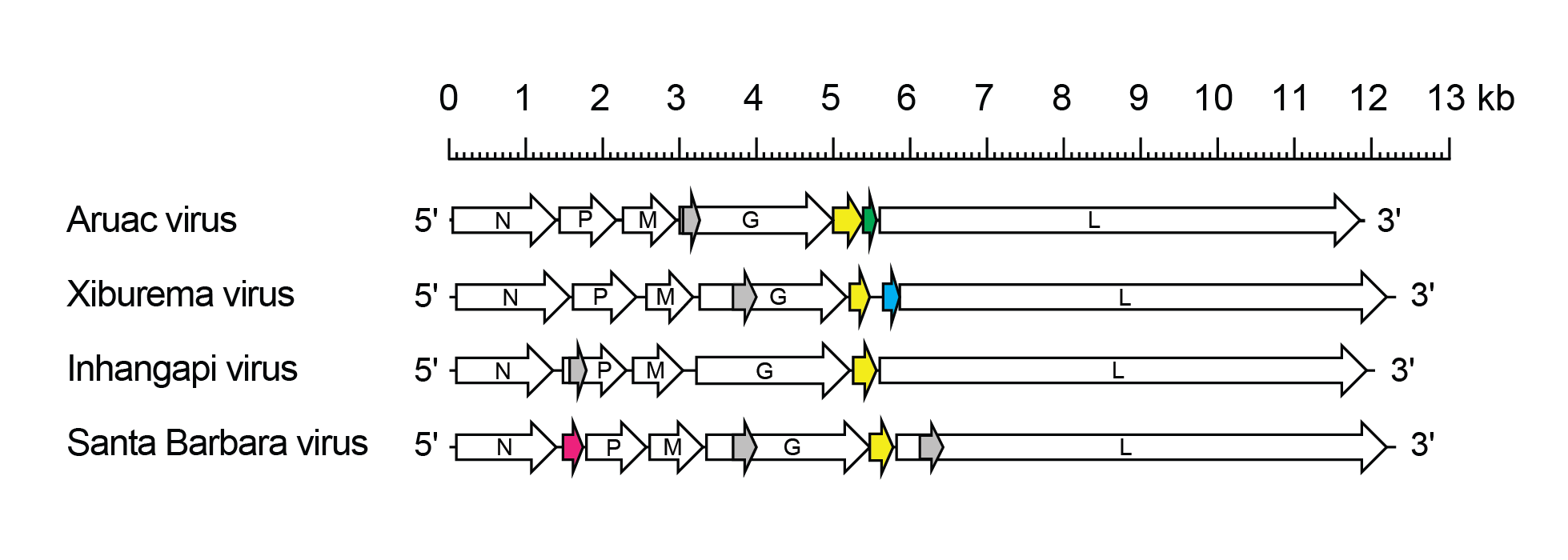 |
| Figure 1.Arurhavirus. Schematic representation of arurhavirus genomes shown in reverse (positive-sense) polarity. Each genome contains long open reading frames (ORFs) in the N, P, M, G and L genes (open arrows) and an ORF encoding a viroporin-like protein (yellow). The Santa Barbara virus U1 ORF (red) and the Xiburema virus U2 ORF (blue) occur in independent transcriptional units (genes). The Aruac virus U1x ORF (green) follows consecutively the U1 ORF within the same transcriptional unit. ORFs >180 nt occur in alternative reading frames within the P, G and L genes (grey) but it is not known if they are expressed. |
Biology
Arurhaviruses have been discovered in forest-dwelling culicine mosquitoes (Diptera: Culicidae) and sandflies (Diptera: Psychodidae) in South America and the Caribbean. ARUV was first isolated from mosquitoes (Trichoprosopon theobaldi) collected from Trinidad, in 1955, and subsequently isolated between 1955 and 1963 from mosquitoes of several other species (Wyeomyia sp., Psorophora ferox, Phoniomyia sp., Culex sp., Sabethes chloropterus) collected from the same region of Trinidad (Spence et al., 1966). XIBV was isolated from mosquitoes (Sabethes intermedius) in Acre State, Brazil, in 1974 (Karabatsos 1985, Wanzeller et al., 2014). INHV was isolated from sandflies (Lutzomyia flaviscutellata) collected in Para State, Brazil, in 1969, and SBAV was detected in sandflies collected in Para State, Brazil, in 2010 (Aitken et al., 1975). Mosquitoes and sandflies of these species feed on birds and mammals, including humans. Neutralising antibodies to INHV have been detected in rodents (Proechimys guyannensis, Hylaeamys megacephalus, Coendou sp.) (Karabatsos 1985) and there is a report of a low prevalence of neutralising antibodies to ARUV in birds (Spence et al., 1968).
Species demarcation criteria
Viruses assigned to different species within the genus Arurhavirus have several of the following characteristics: A) minimum amino acid sequence divergence of 10% in the N proteins; B) minimum amino acid sequence divergence of 10% in the L proteins; C) minimum amino acid sequence divergence of 15% in the G proteins; D) significant differences in genome organisation as evidenced by numbers and locations of ORFs; E) they can be distinguished in virus neutralisation tests; and F) they occupy different ecological niches as evidenced by differences in vertebrate hosts and or arthropod vectors.

