Subfamily: Gammarhabdovirinae
Genus: Novirhabdovirus
Distinguishing features
Viruses assigned to the genus Novirhabdovirus comprise one of the three genera of rhabdoviruses that infect finfish, the others being perhabdoviruses and spriviviruses. Based on well-supported Maximum Likelihood or Maximum Clade Credibility trees inferred from complete L sequences, novirhabdoviruses form a distinct monophyletic group which is phylogenetically distant from spriviviruses and perhabdoviruses, as well as other rhabdoviruses infecting vertebrate hosts. Novirhabdovirus genomes feature an additional gene (NV) located between the G and L genes, encoding a non-structural protein that appears to have a role in blocking the host innate immune response. Although the host range of infectious hematopoietic necrosis virus (IHNV; species Novirhabdovirus salmonid) is limited to salmonid fish, other viruses assigned to the genus have been isolated from fish of numerous diverse taxonomic families.
Virion
Morphology
Virions are bullet-shaped and measure 50–80 nm in diameter and 110–200 nm in length (Kimura et al., 1986, Wolf 1988, Kasornchandra et al., 1992) (Figure 1.Novirhabdovirus). Surface projections are densely dispersed, distinctive spikes which cover the viral surface.
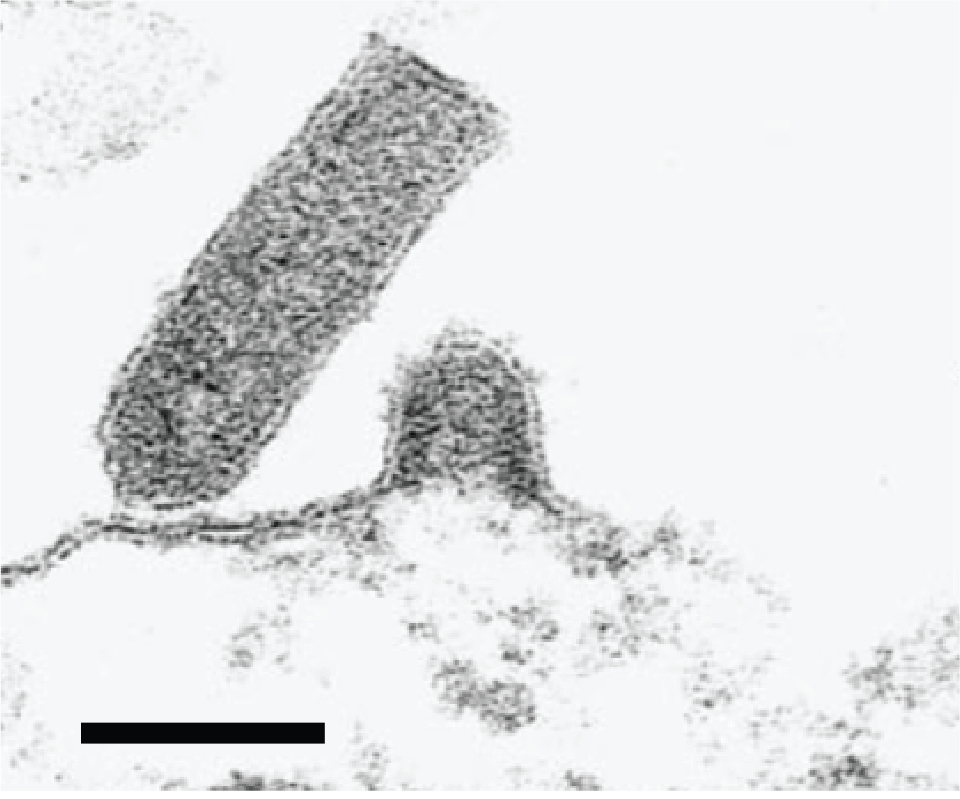 |
| Figure 1.Novirhabdovirus. Electron micrograph showing budding of the fish novirhabdovirus infectious hematopoietic necrosis virus at the plasma membrane and characteristic rhabdovirus bullet-shaped virion structure. Reprinted from Graznow et al. (Figure 2F) (Graznow et al., 1997) with permission from John Wiley and Sons, Publisher. |
Physicochemical and physical properties
The replication temperature range and thermal inactivation temperatures for these viruses are typically lower than those of other rhabdoviruses, due to the aquatic poikilotherm nature of their hosts. Optimum virus replication temperatures range from 15–28 °C, depending roughly on the ambient water temperature in the geographic range of each virus (Wolf 1988, Kurath 2012). IHNV and viral hemorrhagic septicemia virus (VHSV; species Novirhabdovirus piscine) are inactivated above 20 °C.
Nucleic acid
Novirhabdovirus genomes consist of single molecules of negative-sense, single-stranded RNA of approximately 11.1–11.5 kb (Hoffmann et al., 2005, Leong and Kurath 2012).
Proteins
Novirhabdoviruses have five major structural proteins, designated L (150–225 kDa), G (63–80 kDa), N (38–47 kDa), P (22–26 kDa, formerly designated M1), and M (17–22 kDa, formerly designated M2) (Lenoir and de Kinkelin 1975, McAllister and Wagner 1975, Leong and Kurath 2012). In addition to the structural proteins, novirhabdovirus genomes encode a small, sixth, non-virion protein designated NV (12–14 kDa), which is expressed at variable levels in infected cells but is not detectable in purified virions (Kurath et al., 1985, Kurath and Leong 1985, Schutze et al., 1996). The NV ORF is encoded on an independent transcription unit between the G gene and L gene. The NV gene and ORF are absolutely preserved in numerous diverse viruses assigned to the four novirhabdovirus species, but the NV protein sequences are significantly less conserved between viruses in different species than sequences of the other structural proteins; there is no significant amino acid sequence similarity between the NV proteins of viruses of different species (Kurath et al., 1997). The NV protein has been shown to localize to the nucleus in infected cells (Choi et al., 2011), and it has also been shown to inhibit apoptosis and interfere with the host innate immune response (Ammayappan and Vakharia 2011, Kim and Kim 2013, Biacchesi et al., 2017). Studies with NV gene deletion mutants generated by reverse genetics have shown that the NV protein is not essential for virus viability but it is required for efficient virus replication (Biacchesi et al., 2000, Johnson et al., 2000). Recombinant virus studies are inconsistent in that the NV gene appears to be required for pathogenicity in IHNV and VHSV but not snakehead rhabdovirus (SHRV; species Novirhabdovirus snakehead) (Johnson et al., 2000, Alonso et al., 2004, Thoulouze et al., 2004, Ammayappan et al., 2011).
Genome organisation and replication
The genomic RNA contains six genes in the order 3′-N-P-M-G-NV-L-5′ (Kurath and Leong 1985, Schutze et al., 1995, Johnson et al., 2000, Kim et al., 2005) (Figure 2.Novirhabdovirus). A leader region of approximately 60 nt precedes the transcription initiation of the N gene and a trailer of approximately 100 nt follows the transcription termination of the L gene. Genes begin with the conserved putative transcription initiation signal 3′-CCRWG (vRNA sense, most often 3′-CCGUG), which has been experimentally confirmed as the 5′-terminus of viral mRNA for various genes of IHNV, VHSV, and SHRV. All genes end with the conserved transcription termination/polyadenylation signal 3′-UCURUC(U)7; non-transcribed intergenic regions are single nucleotides, G or A (vRNA sense).
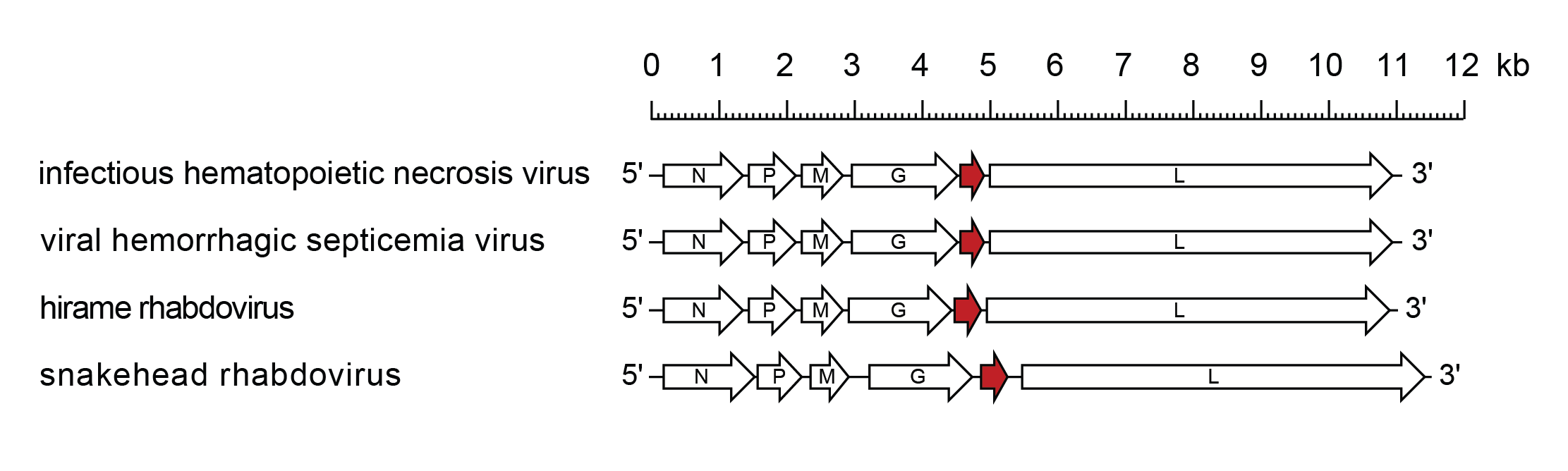 |
| Figure 2.Novirhabdovirus. Schematic representation of novirhabdovirus genomes shown in reverse (positive-sense) polarity. N, P, M, G and L represent ORFs encoding the structural proteins. ORF NV (U1), encoding a protein involved in pathogenicity and blocking host innate immunity, is highlighted (red brown). |
Biology
Novirhabdoviruses infect fish of numerous species. The natural host range of individual viruses varies in breadth, with IHNV limited to salmonid fish, while VHSV infects over 80 host species from a wide range of fish families as diverse as salmonids (Salmonidae) and herring (Clupeidae) (Bootland and Leong 1999, Office International des Epizooties 2009). In nature and in artificial environments, novirhabdoviruses can be transmitted horizontally by waterborne virus shed from infected fish (Wolf 1988, Bootland and Leong 1999, Smail 1999). Egg-associated transmission has also been clearly demonstrated in several cases in which the spread of virus to new geographic regions has occurred through transport of contaminated fish eggs; however, egg disinfection with iodophore treatment is very effective, suggesting virus infection is typically not within the egg (Bootland and Leong 1999, Dixon et al., 2016). It is increasingly apparent that wild fish can serve as novirhabdovirus reservoirs (Skall et al., 2005, Kurath and Winton 2011, Dixon et al., 2016). The existence of invertebrate reservoirs or vectors of virus has been postulated but their ecological importance is uncertain (Jakob et al., 2011, Dixon et al., 2016). Similarly, the potential for a carrier state in survivors of IHNV or VHSV infections has been demonstrated but the frequency and significance of this phenomenon is not well understood (Bootland and Leong 1999, Dixon et al., 2016).
The geographic distribution of novirhabdoviruses is broad. IHNV is enzootic to western North America but inadvertent transport of the virus with contaminated eggs and fish has resulted in spread and establishment of IHNV in Europe and Asia (including China and Iran) (Wolf 1988, Kurath 2012, Jia et al., 2014, Adel et al., 2016). VHSV occurs in an extensive reservoir of diverse wild fish species in marine waters in the northern Atlantic and Pacific Oceans, as well as the freshwater Great Lakes in North America (Smail 1999, Skall et al., 2005, Faisal et al., 2012, Garver et al., 2013). VHSV is also enzootic in cultured rainbow trout in much of Western Europe and in cultured flounder in Asia (Skall et al., 2005, Nishizawa et al., 2006). Hirame rhabdovirus (HIRRV; species Novirhabdovirus hirame) is detected predominantly in Asia (Kimura et al., 1986, Kim and Oh 2015), although it has been reported recently in Poland (Borzym et al., 2014). SHRV has only been reported in South-East Asia (Kasornchandra et al., 1992).
Novirhabdoviruses cause disease in cultured fish hosts, resulting in significant economic losses to aquaculture industries. Both IHNV and VHSV have been well documented as major pathogens of cultured salmonids since the 1950s, often resulting in losses of 50–100% (Bootland and Leong 1999, Smail 1999). Among free-ranging fish, IHNV epizootics have been reported in wild salmonids and VHSV epizootics have occurred in both marine and freshwater fish of diverse species (Wolf 1988, Faisal et al., 2012, Garver et al., 2013). IHNV and VHSV both cause haemorrhagic diseases, with petechial haemorrhages evident both externally and internally. Major degenerative changes and necrosis in the kidneys and haematopoietic tissue are evident upon histopathological examination and are believed to be the actual cause of mortality (Bootland and Leong 1999, Smail 1999).
Antigenicity
For IHNV and VHSV, G has been identified as the major antigenic protein. Fish initially mount a strong interferon-based innate immune response to infection, followed by both humoral and cellular adaptive immune responses (Purcell et al., 2012). In western blots, fish antibodies to multiple viral proteins are detected, but vaccine studies have shown that antibodies to G are necessary and sufficient alone to provide protective immunity (Engelking and Leong 1989, Corbeil et al., 1999). Viruses assigned to different species within the genus have been distinguished serologically based on cross-neutralisation with polyclonal rabbit antisera. In general, virus strains assigned to a species are neutralised by a single polyclonal antiserum, but with varying efficiencies. Thus, IHNV and HIRRV each comprise single serotypes and VHSV has one major serotype with a small number of associated strains. Viruses assigned to different species do not show cross-neutralisation but, in some cases, there is a low level of cross-reaction with specific proteins in western blot analyses.
Species demarcation criteria
Novirhabdovirus species were originally defined on the basis of lack of serological cross-reactivity which is supported by phylogenetic analyses of various genes or complete genomes. Host and geographic range are additional characteristics used to distinguish species within the genus. All novirhabdoviruses share the same genome architecture with six homologous genes in the same order so this is not a distinguishing trait.
Viruses assigned to different species within the genus Novirhabdovirus have some of the following characteristics: A) minimum of 31% nucleotide divergence and 26% amino acid divergence in the G gene and G protein, respectively; B) phylogenetic structure as distinct monophyletic clades in phylogenetic analyses with various genome regions, obtained with various evolutionary models; C) distinguished by serological tests; and D) occupy different ecological niches as indicated by non-overlapping or partially overlapping host ranges, and non-overlapping or partially overlapping geographic ranges. All members currently meet criteria A, B, and C, and some members differ from others by D.
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation |
| Carpione rhabdovirus | LC630942 | CAPRV |
Virus names and virus abbreviations are not official ICTV designations.
Genera unassigned to a subfamily

